Intest Res.
2024 Apr;22(2):131-151. 10.5217/ir.2023.00115.
Pathogenesis and biomarkers of colorectal cancer by epigenetic alteration
- Affiliations
-
- 1Division of Gastroenterology, Department of Internal Medicine, Hallym University Kangnam Sacred Heart Hospital, Hallym University College of Medicine, Seoul, Korea
- 2Division of Gastroenterology, Department of Internal Medicine, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2554658
- DOI: http://doi.org/10.5217/ir.2023.00115
Abstract
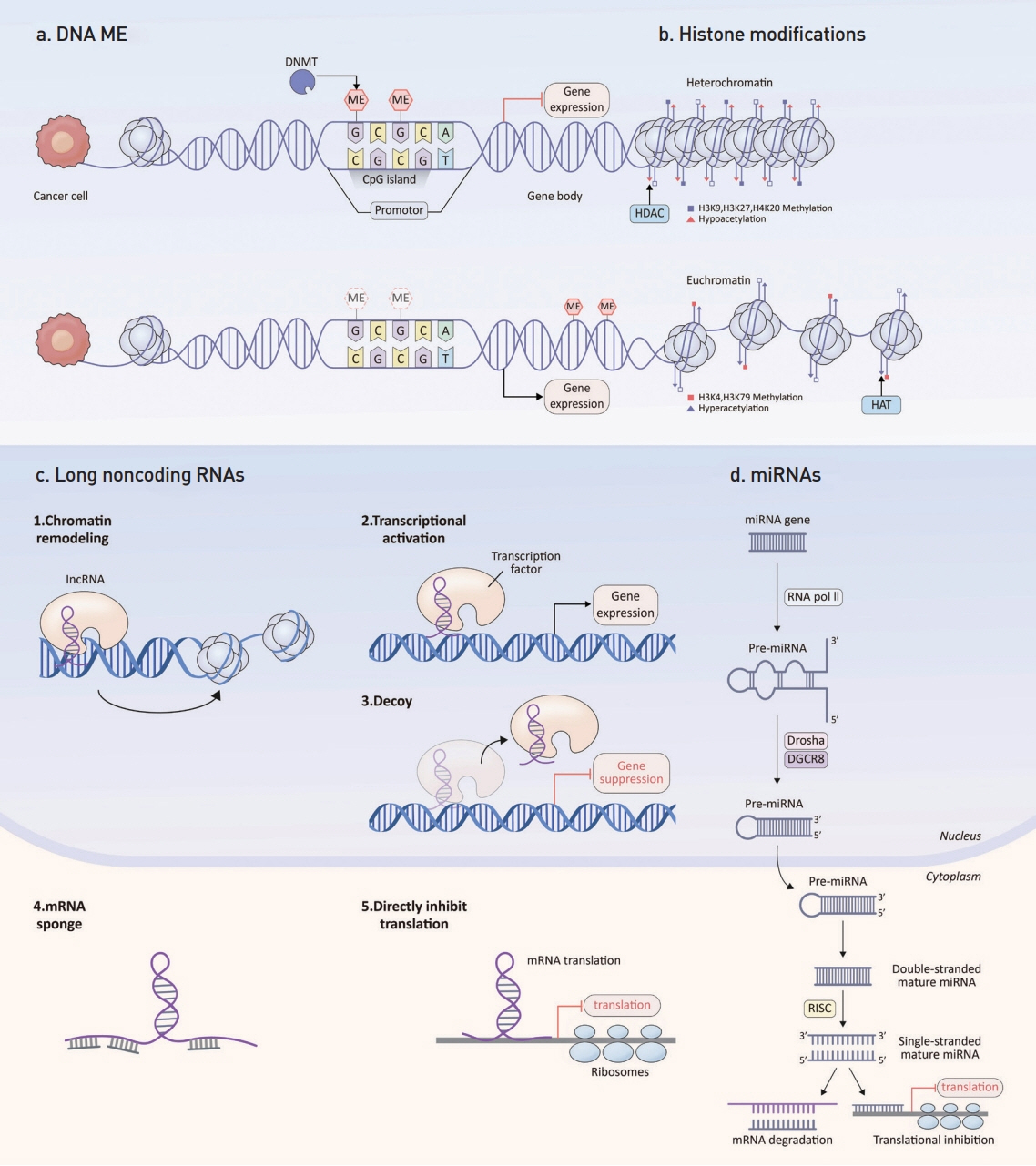
- Colorectal cancer (CRC) ranks third in cancer incidence and stands as the second leading cause of cancer-related deaths globally. CRC tumorigenesis results from a cumulative set of genetic and epigenetic alterations, disrupting cancer-regulatory processes like cell proliferation, metabolism, angiogenesis, cell death, invasion, and metastasis. Key epigenetic modifications observed in cancers encompass abnormal DNA methylation, atypical histone modifications, and irregularities in noncoding RNAs, such as microRNAs and long noncoding RNAs. The advancement in genomic technologies has positioned these genetic and epigenetic shifts as potential clinical biomarkers for CRC patients. This review concisely covers the fundamental principles of CRC-associated epigenetic changes, and examines in detail their emerging role as biomarkers for early detection, prognosis, and treatment response prediction.
Keyword
Figure
Reference
-
1. Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015; 149:1204–1225.
Article2. Estécio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007; 2:e399.
Article3. Inamura K, Yamauchi M, Nishihara R, et al. Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis. J Natl Cancer Inst. 2014; 106–dju195.
Article4. Grady WM, Willis J, Guilford PJ, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffusee gastric cancer. Nat Genet. 2000; 26:16–17.
Article5. Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008; 122:2767–2773.
Article6. Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Sci Rep. 2017; 7:3032.
Article7. Wu D, Zhou G, Jin P, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016; 18:535–545.
Article8. Chi P, Allis CD, Wang GG. Covalent histone modifications: miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010; 10:457–469.
Article9. Hanigan CL, Van Engeland M, De Bruine AP, et al. An inactivating mutation in HDAC2 leads to dysregulation of apoptosis mediated by APAF1. Gastroenterology. 2008; 135:1654–1664.
Article10. Gargalionis AN, Piperi C, Adamopoulos C, Papavassiliou AG. Histone modifications as a pathogenic mechanism of colorectal tumorigenesis. Int J Biochem Cell Biol. 2012; 44:1276–1289.
Article11. Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998; 12:599–606.12. Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016; 8–a019521.
Article13. Huang T, Lin C, Zhong LL, et al. Targeting histone methylation for colorectal cancer. Therap Adv Gastroenterol. 2017; 10:114–131.
Article14. Salz T, Li G, Kaye F, Zhou L, Qiu Y, Huang S. hSETD1A regulates Wnt target genes and controls tumor growth of colorectal cancer cells. Cancer Res. 2014; 74:775–786.
Article15. Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012; 13:343–357.
Article16. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011; 12:861–874.
Article17. He N, Song L, Kang Q, et al. The pathological features of colorectal cancer determine the detection performance on blood ctDNA. Technol Cancer Res Treat. 2018; 17:1533033818791794.
Article18. Fu B, Yan P, Zhang S, et al. Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers. 2018; 2018:6437104.
Article19. Song L, Peng X, Li Y, et al. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics. 2017; 9:599–610.
Article20. Tänzer M, Balluff B, Distler J, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010; 5:e9061.
Article21. Grützmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by Septin 9 DNA methylation assay. PLoS One. 2008; 3:e3759.
Article22. Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Invest Med. 2011; 34–E88-E95.
Article23. Barták BK, Kalmár A, Péterfia B, et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics. 2017; 12:751–763.
Article24. Oh T, Kim N, Moon Y, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013; 15:498–507.
Article25. Lee BB, Lee EJ, Jung EH, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009; 15:6185–6191.
Article26. Cassinotti E, Melson J, Liggett T, et al. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int J Cancer. 2012; 131:1153–1157.
Article27. He Q, Chen HY, Bai EQ, et al. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet. 2010; 202:1–10.
Article28. Rasmussen SL, Krarup HB, Sunesen KG, et al. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS One. 2017; 12:e0180809.
Article29. Zhao G, Li H, Yang Z, et al. Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 2019; 8:5619–5628.
Article30. Chen Y, Wang Z, Zhao G, et al. Performance of a novel bloodbased early colorectal cancer screening assay in remaining serum after the blood biochemical test. Dis Markers. 2019; 2019:5232780.
Article31. Pedersen SK, Symonds EL, Baker RT, et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015; 15:654.
Article32. Jensen SØ, Øgaard N, Ørntoft MW, et al. Novel DNA methylation biomarkers show high sensitivity and specificity for bloodbased detection of colorectal cancer-a clinical biomarker discovery and validation study. Clin Epigenetics. 2019; 11:158.33. Ebert MP, Model F, Mooney S, et al. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006; 131:1418–1430.
Article34. Zou HZ, Yu BM, Wang ZW, et al. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clin Cancer Res. 2002; 8:188–191.35. Wallner M, Herbst A, Behrens A, et al. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res. 2006; 12:7347–7352.36. Leung WK, To KF, Man EP, et al. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am J Gastroenterol. 2005; 100:2274–2279.37. Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001; 61:900–902.
Article38. Herbst A, Rahmig K, Stieber P, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011; 106:1110–1118.39. Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008; 54:414–423.
Article40. Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009; 27:858–863.41. Xiao W, Zhao H, Dong W, et al. Quantitative detection of methylated NDRG4 gene as a candidate biomarker for diagnosis of colorectal cancer. Oncol Lett. 2015; 9:1383–1387.42. Esteller M, González S, Risques RA, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol. 2001; 19:299–304.43. Nakayama H, Hibi K, Taguchi M, et al. Molecular detection of p16 promoter methylation in the serum of colorectal cancer patients. Cancer Lett. 2002; 188:115–119.44. Nakayama H, Hibi K, Takase T, et al. Molecular detection of p16 promoter methylation in the serum of recurrent colorectal cancer patients. Int J Cancer. 2003; 105:491–493.45. Lecomte T, Berger A, Zinzindohoué F, et al. Detection of freecirculating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002; 100:542–548.46. Xing XB, Cai WB, Luo L, Liu LS, Shi HJ, Chen MH. The prognostic value of p16 hypermethylation in cancer: a meta-analysis. PLoS One. 2013; 8:e66587.47. Mitomi H, Fukui N, Tanaka N, et al. Aberrant p16((INK4a)) methylation is a frequent event in colorectal cancers: prognostic value and relation to mRNA expression and immunoreactivity. J Cancer Res Clin Oncol. 2010; 136:323–331.48. Shen L, Catalano PJ, Benson AB, O’Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007; 13:6093–6098.49. Kim JC, Choi JS, Roh SA, Cho DH, Kim TW, Kim YS. Promoter methylation of specific genes is associated with the phenotype and progression of colorectal adenocarcinomas. Ann Surg Oncol. 2010; 17:1767–1776.50. Antelo M, Balaguer F, Shia J, et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS One. 2012; 7:e45357.
Article51. Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008; 100:1734–1738.52. Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011; 117:1847–1854.
Article53. Rhee YY, Kim MJ, Bae JM, et al. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol. 2012; 19:3441–3448.54. Kawakami K, Matsunoki A, Kaneko M, Saito K, Watanabe G, Minamoto T. Long interspersed nuclear element-1 hypomethylation is a potential biomarker for the prediction of response to oral fluoropyrimidines in microsatellite stable and CpG island methylator phenotype-negative colorectal cancer. Cancer Sci. 2011; 102:166–174.55. Philipp AB, Nagel D, Stieber P, et al. Circulating cell-free methylated DNA and lactate dehydrogenase release in colorectal cancer. BMC Cancer. 2014; 14:245.
Article56. Philipp AB, Stieber P, Nagel D, et al. Prognostic role of methylated free circulating DNA in colorectal cancer. Int J Cancer. 2012; 131:2308–2319.57. Herbst A, Vdovin N, Gacesa S, et al. Methylated free-circulating HPP1 DNA is an early response marker in patients with metastatic colorectal cancer. Int J Cancer. 2017; 140:2134–2144.58. Yi JM, Dhir M, Van Neste L, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011; 17:1535–1545.
Article59. Nagasaka T, Sharp GB, Notohara K, et al. Hypermethylation of O6-methylguanine-DNA methyltransferase promoter may predict nonrecurrence after chemotherapy in colorectal cancer cases. Clin Cancer Res. 2003; 9:5306–5312.60. Amatu A, Sartore-Bianchi A, Moutinho C, et al. Promoter CpG island hypermethylation of the DNA repair enzyme MGMT predicts clinical response to dacarbazine in a phase II study for metastatic colorectal cancer. Clin Cancer Res. 2013; 19:2265–2272.
Article61. Cleven AH, Derks S, Draht MX, et al. CHFR promoter methylation indicates poor prognosis in stage II microsatellite stable colorectal cancer. Clin Cancer Res. 2014; 20:3261–3271.62. Goltz D, Gevensleben H, Dietrich J, Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2016; 6:e1257454.63. Iwata N, Ishikawa T, Okazaki S, et al. Clinical significance of methylation and reduced expression of the quaking gene in colorectal cancer. Anticancer Res. 2017; 37:489–498.64. Draht MX, Smits KM, Tournier B, et al. Promoter CpG island methylation of RET predicts poor prognosis in stage II colorectal cancer patients. Mol Oncol. 2014; 8:679–688.65. Liu Y, Chew MH, Tham CK, Tang CL, Ong SY, Zhao Y. Methylation of serum SST gene is an independent prognostic marker in colorectal cancer. Am J Cancer Res. 2016; 6:2098–2108.66. Zhang ZM, Wang Y, Huang R, et al. TFAP2E hypermethylation was associated with survival advantage in patients with colorectal cancer. J Cancer Res Clin Oncol. 2014; 140:2119–2127.67. Moutinho C, Martinez-Cardús A, Santos C, et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst. 2014; 106–djt322.68. Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH, Park DI. Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Dis Colon Rectum. 2009; 52:1452–1459.
Article69. Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005; 97:1124–1132.
Article70. Zou H, Harrington JJ, Shire AM, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007; 16:2686–2696.
Article71. Ned RM, Melillo S, Marrone M. Fecal DNA testing for colorectal cancer screening: the ColoSure™ test. PLoS Curr. 2011; 3:RR–N1220.
Article72. Melotte V, Lentjes MH, van den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009; 101:916–927.
Article73. Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014; 370:1287–1297.
Article74. Zhang W, Yang C, Wang S, et al. SDC2 and TFPI2 methylation in stool samples as an integrated biomarker for early detection of colorectal cancer. Cancer Manag Res. 2021; 13:3601–3617.
Article75. Park SK, Baek HL, Yu J, et al. Is methylation analysis of SFRP2, TFPI2, NDRG4, and BMP3 promoters suitable for colorectal cancer screening in the Korean population? Intest Res. 2017; 15:495–501.76. Lu H, Huang S, Zhang X, et al. DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA. Oncol Lett. 2014; 8:1751–1756.77. Amiot A, Mansour H, Baumgaertner I, et al. The detection of the methylated Wif-1 gene is more accurate than a fecal occult blood test for colorectal cancer screening. PLoS One. 2014; 9:e99233.
Article78. Abbaszadegan MR, Tavasoli A, Velayati A, et al. Stool-based DNA testing, a new noninvasive method for colorectal cancer screening, the first report from Iran. World J Gastroenterol. 2007; 13:1528–1533.
Article79. Petko Z, Ghiassi M, Shuber A, et al. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005; 11:1203–1209.
Article80. Chang E, Park DI, Kim YJ, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepatogastroenterology. 2010; 57:720–727.81. Hellebrekers DM, Lentjes MH, van den Bosch SM, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res. 2009; 15:3990–3997.
Article82. Ausch C, Kim YH, Tsuchiya KD, et al. Comparative analysis of PCR-based biomarker assay methods for colorectal polyp detection from fecal DNA. Clin Chem. 2009; 55:1559–1563.
Article83. Huang ZH, Li LH, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol. 2007; 13:950–954.
Article84. Bosch LJ, Oort FA, Neerincx M, et al. DNA methylation of phosphatase and actin regulator 3 detects colorectal cancer in stool and complements FIT. Cancer Prev Res (Phila). 2012; 5:464–472.
Article85. Nagasaka T, Tanaka N, Cullings HM, et al. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009; 101:1244–1258.
Article86. Carmona FJ, Azuara D, Berenguer-Llergo A, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res (Phila). 2013; 6:656–665.
Article87. Zhang H, Zhu YQ, Wu YQ, Zhang P, Qi J. Detection of promoter hypermethylation of Wnt antagonist genes in fecal samples for diagnosis of early colorectal cancer. World J Gastroenterol. 2014; 20:6329–6335.
Article88. Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009; 10:295–304.
Article89. Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015; 7:1051–1073.
Article90. Kouzarides T. SnapShot: histone-modifying enzymes. Cell. 2007; 131:822.
Article91. Nakazawa T, Kondo T, Ma D, et al. Global histone modification of histone H3 in colorectal cancer and its precursor lesions. Hum Pathol. 2012; 43:834–842.
Article92. Karczmarski J, Rubel T, Paziewska A, et al. Histone H3 lysine 27 acetylation is altered in colon cancer. Clin Proteomics. 2014; 11:24.
Article93. Ashktorab H, Belgrave K, Hosseinkhah F, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci. 2009; 54:2109–2117.
Article94. Gezer U, Ustek D, Yörüker EE, et al. Characterization of H3K9me3- and H4K20me3-associated circulating nucleosomal DNA by high-throughput sequencing in colorectal cancer. Tumour Biol. 2013; 34:329–336.
Article95. Gezer U, Yörüker EE, Keskin M, Kulle CB, Dharuman Y, Holdenrieder S. Histone methylation marks on circulating nucleosomes as novel blood-based biomarker in colorectal cancer. Int J Mol Sci. 2015; 16:29654–29662.
Article96. Tamagawa H, Oshima T, Shiozawa M, et al. The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol Rep. 2012; 27:637–642.
Article97. Tamagawa H, Oshima T, Numata M, et al. Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur J Surg Oncol. 2013; 39:655–661.
Article98. Benard A, Goossens-Beumer IJ, van Hoesel AQ, et al. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer. 2014; 14:531.
Article99. Yokoyama Y, Hieda M, Nishioka Y, et al. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 2013; 104:889–895.
Article100. Goossens-Beumer IJ, Benard A, van Hoesel AQ, et al. Agedependent clinical prognostic value of histone modifications in colorectal cancer. Transl Res. 2015; 165:578–588.
Article101. Benard A, Goossens-Beumer IJ, van Hoesel AQ, et al. Prognostic value of polycomb proteins EZH2, BMI1 and SUZ12 and histone modification H3K27me3 in colorectal cancer. PLoS One. 2014; 9:e108265.
Article102. Benard A, Goossens-Beumer IJ, van Hoesel AQ, et al. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology. 2015; 66:270–282.
Article103. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297.104. Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer. 2006; 6:259–269.105. Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003; 1:882–891.106. Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011; 20:1272–1286.
Article107. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011; 39:7223–7233.
Article108. Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008; 299:425–436.
Article109. Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013; 105:849–859.
Article110. Liu T, Liu D, Guan S, Dong M. Diagnostic role of circulating MiR-21 in colorectal cancer: a update meta-analysis. Ann Med. 2021; 53:87–102.
Article111. Jenike AE, Halushka MK. miR-21: a non-specific biomarker of all maladies. Biomark Res. 2021; 9:18.
Article112. Santos DAR, Gaiteiro C, Santos M, Santos L, Dinis-Ribeiro M, Lima L. MicroRNA biomarkers as promising tools for early colorectal cancer screening: a comprehensive review. Int J Mol Sci. 2023; 24:11023.
Article113. Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013; 20:1603–1614.
Article114. Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009; 58:1375–1381.
Article115. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010; 127:118–126.
Article116. Yang X, Zeng Z, Hou Y, et al. MicroRNA-92a as a potential biomarker in diagnosis of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014; 9:e88745.
Article117. Moloudizargari M, Rahmani J, Asghari MH, Goel A. The prognostic role of miR-31 in colorectal cancer: the results of a meta-analysis of 4720 patients. Epigenomics. 2022; 14:101–112.
Article118. Moody L, Dvoretskiy S, An R, Mantha S, Pan YX. The efficacy of miR-20a as a diagnostic and prognostic biomarker for colorectal cancer: a systematic review and meta-analysis. Cancers (Basel). 2019; 11:1111.
Article119. Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and metaanalysis. Br J Cancer. 2017; 116:762–774.
Article120. Zuo Z, Jiang Y, Zeng S, et al. The value of microRNAs as the novel biomarkers for colorectal cancer diagnosis: a meta-analysis. Pathol Res Pract. 2020; 216:153130.
Article121. Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020; 17:111–130.
Article122. Liu HN, Liu TT, Wu H, et al. Serum microRNA signatures and metabolomics have high diagnostic value in colorectal cancer using two novel methods. Cancer Sci. 2018; 109:1185–1194.123. Liu J, Chen B, Yang M, et al. A three-plasma miRNA panel predicts the risk of colorectal cancer: a community-based nested case‒control study. Sci Rep. 2023; 13:4196.
Article124. Basati G, Razavi AE, Pakzad I, Malayeri FA. Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol. 2016; 37:1781–1788.
Article125. Peng J, Xie Z, Cheng L, et al. Paired design study by real-time PCR: miR-378* and miR-145 are potent early diagnostic biomarkers of human colorectal cancer. BMC Cancer. 2015; 15:158.
Article126. Sur D, Advani S, Braithwaite D. MicroRNA panels as diagnostic biomarkers for colorectal cancer: a systematic review and meta-analysis. Front Med (Lausanne). 2022; 9:915226.
Article127. Nakamura K, Hernández G, Sharma GG, et al. A liquid biopsy signature for the detection of patients with early-onset colorectal cancer. Gastroenterology. 2022; 163:1242–1251.
Article128. Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer: implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018; 15:617–638.
Article129. Mezher M, Abdallah S, Ashekyan O, et al. Insights on the biomarker potential of exosomal non-coding RNAs in colorectal cancer: an in silico characterization of related exosomal lncRNA/circRNA-miRNA-target axis. Cells. 2023; 12:1081.
Article130. Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014; 9:e92921.
Article131. Maminezhad H, Ghanadian S, Pakravan K, et al. A panel of six-circulating miRNA signature in serum and its potential diagnostic value in colorectal cancer. Life Sci. 2020; 258:118226.
Article132. Wang CJ, Zhou ZG, Wang L, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009; 26:27–34.
Article133. Kjersem JB, Ikdahl T, Lingjaerde OC, Guren T, Tveit KM, Kure EH. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol. 2014; 8:59–67.
Article134. Gherman A, Balacescu L, Popa C, et al. Baseline expression of exosomal miR-92a-3p and miR-221-3p could predict the response to first-line chemotherapy and survival in metastatic colorectal cancer. Int J Mol Sci. 2023; 24:10622.
Article135. Wu CW, Ng SS, Dong YJ, et al. Detection of miR-92a and miR21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012; 61:739–745.
Article136. Kalimutho M, Del Vecchio Blanco G, Di Cecilia S, et al. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011; 46:1391–1402.
Article137. Ahmed FE, Jeffries CD, Vos PW, et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009; 6:281–295.138. Koga Y, Yasunaga M, Takahashi A, et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila). 2010; 3:1435–1442.
Article139. Choi HH, Cho YS, Choi JH, Kim HK, Kim SS, Chae HS. Stoolbased miR-92a and miR-144* as noninvasive biomarkers for colorectal cancer screening. Oncology. 2019; 97:173–179.
Article140. Duran-Sanchon S, Moreno L, Augé JM, et al. Identification and validation of microRNA profiles in fecal samples for detection of colorectal cancer. Gastroenterology. 2020; 158:947–957.
Article141. Zhu Y, Xu A, Li J, et al. Fecal miR-29a and miR-224 as the noninvasive biomarkers for colorectal cancer. Cancer Biomark. 2016; 16:259–264.
Article142. Duran-Sanchon S, Moreno L, Gómez-Matas J, et al. Fecal microRNA-based algorithm increases effectiveness of fecal immunochemical test-based screening for colorectal cancer. Clin Gastroenterol Hepatol. 2021; 19:323–330.
Article143. Zhao Z, Zhu A, Bhardwaj M, Schrotz-King P, Brenner H. Fecal microRNAs, fecal microRNA panels, or combinations of fecal microRNAs with fecal hemoglobin for early detection of colorectal cancer and its precursors: a systematic review. Cancers (Basel). 2021; 14:65.
Article144. Pardini B, Ferrero G, Tarallo S, et al. A fecal microRNA signature by small RNA sequencing accurately distinguishes colorectal cancers: results from a multicenter study. Gastroenterology. 2023; 165:582–599.
Article145. Koga Y, Yamazaki N, Yamamoto Y, et al. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev. 2013; 22:1844–1852.
Article146. Yang Y, Yan X, Li X, Ma Y, Goel A. Long non-coding RNAs in colorectal cancer: novel oncogenic mechanisms and promising clinical applications. Cancer Lett. 2021; 504:67–80.
Article147. Lulli M, Napoli C, Landini I, Mini E, Lapucci A. Role of noncoding RNAs in colorectal cancer: focus on long non-coding RNAs. Int J Mol Sci. 2022; 23:13431.
Article148. Wu Y, Xu X. Long non-coding RNA signature in colorectal cancer: research progression and clinical application. Cancer Cell Int. 2023; 23:28.
Article149. Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011; 71:6320–6326.
Article150. Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014; 35:1510–1515.
Article151. Alaiyan B, Ilyayev N, Stojadinovic A, et al. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013; 13:196.
Article152. Ozawa T, Matsuyama T, Toiyama Y, et al. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017; 28:1882–1888.
Article153. Zhao W, Song M, Zhang J, Kuerban M, Wang H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol. 2015; 8:14131–14140.154. Hu Z, Wu J, Tan S, et al. Diagnostic value of long noncoding RNA LINC01485 in patients with colorectal cancer. Clin Biochem. 2022; 102:34–43.
Article155. Tao K, Yang J, Hu Y, et al. Clinical significance of urothelial carcinoma associated 1 in colon cancer. Int J Clin Exp Med. 2015; 8:21854–21860.156. Pichler M, Rodriguez-Aguayo C, Nam SY, et al. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut. 2020; 69:1818–1831.
Article157. Xu J, Meng Q, Li X, et al. Long noncoding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Cancer Res. 2019; 79:4882–4895.
Article158. Damas ND, Marcatti M, Côme C, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016; 7:13875.
Article159. Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018; 11:113.
Article160. Shi J, Li X, Zhang F, et al. Circulating lncRNAs associated with occurrence of colorectal cancer progression. Am J Cancer Res. 2015; 5:2258–2265.161. Gharib E, Nazemalhosseini-Mojarad E, Baghdar K, et al. Identification of a stool long non-coding RNAs panel as a potential biomarker for early detection of colorectal cancer. J Clin Lab Anal. 2021; 35:e23601.
Article162. Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041.
Article163. Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014; 7:3174–3181.164. Siddique H, Al-Ghafari A, Choudhry H, et al. Long noncoding RNAs as prognostic markers for colorectal cancer in Saudi patients. Genet Test Mol Biomarkers. 2019; 23:509–514.
Article165. Chen S, Zhang C, Feng M. Prognostic value of lncRNA HOTAIR in colorectal cancer: a meta-analysis. Open Med (Wars). 2020; 15:76–83.166. Ohtsuka M, Ling H, Ivan C, et al. H19 noncoding RNA, an independent prognostic factor, regulates essential Rb-E2F and CDK8-β-catenin signaling in colorectal cancer. EBioMedicine. 2016; 13:113–124.
Article167. Ren YK, Xiao Y, Wan XB, et al. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015; 8:11458–11463.168. Wu Y, Yang X, Chen Z, et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019; 18:87.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic and epigenetic alterations of colorectal cancer
- Understanding of molecular pathogenesis and genetic markers in colorectal cancer
- Epigenetic Alterations in Inflammatory Bowel Disease and Cancer
- Stool Based DNA Biomarkers for Colorectal Cancer Diagnosis
- Clinical Application of Genetics in Management of Colorectal Cancer