Clin Exp Otorhinolaryngol.
2023 May;16(2):115-124. 10.21053/ceo.2022.01235.
Identification and Characterization of mRNA and lncRNA Expression Profiles in Age-Related Hearing Loss
- Affiliations
-
- 1Department of Neurology, Chonnam National University Hospital, Gwangju, Korea
- 2Department of Biochemistry, Chonnam National University Medical School, Gwangju, Korea
- 3Department of Otolaryngology-Head and Neck Surgery, Chonnam National University Hospital, Chonnam National University Medical School, Gwangju, Korea
- 4Department of Neurology, Chonnam National University Medical School, Gwangju, Korea
- KMID: 2542352
- DOI: http://doi.org/10.21053/ceo.2022.01235
Abstract
Objectives
. Age-related hearing loss (ARHL), or presbycusis, is caused by disorders of sensory hair cells and auditory neurons. Many studies have suggested that the accumulation of mitochondrial DNA damage, the production of reactive oxygen species, noise, inflammation, and decreased antioxidant function are associated with subsequent cochlear senescence in response to aging stress. Long non-coding RNA (lncRNA) has been reported to play important roles in various diseases. However, the function of lncRNA in ARHL remains unclear. In this study, we analyzed the common expression profiles of messenger RNA (mRNA) and lncRNA through ARHL-related RNA-sequencing datasets.
Methods
. We selected and downloaded three different sets of RNA-sequencing data for ARHL. We performed differential expression analysis to find common mRNA and lncRNA profiles in the cochleae of aged mice compared to young mice. Gene Ontology (GO) analysis was used for functional exploration. Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed to validate mRNAs and lncRNAs. In addition, we performed trans target prediction analysis with differentially expressed mRNAs and lncRNAs to understand the function of these mRNAs and lncRNAs in ARHL.
Results
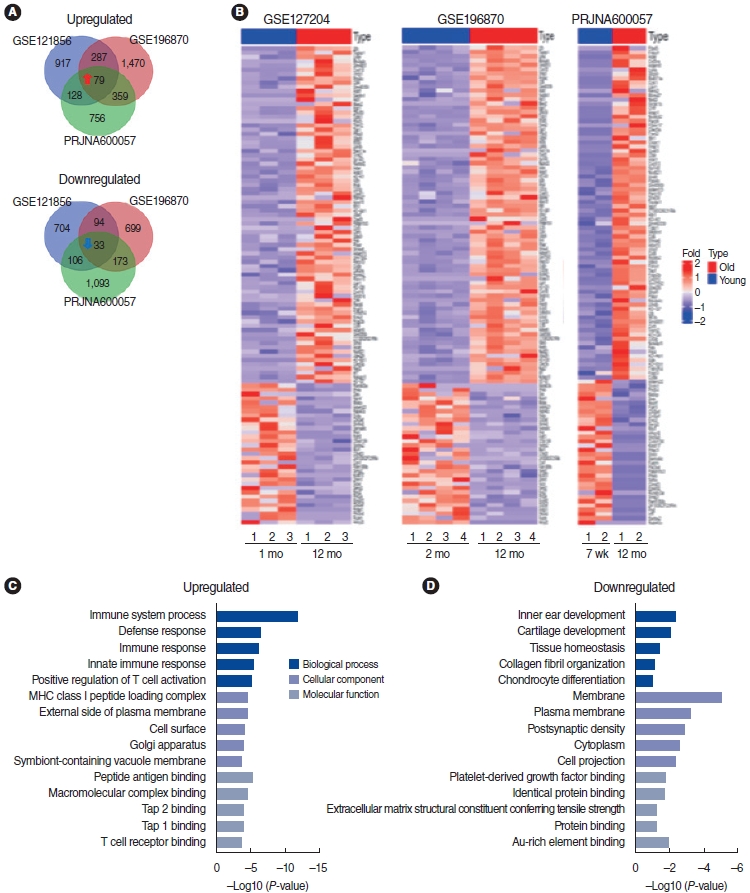
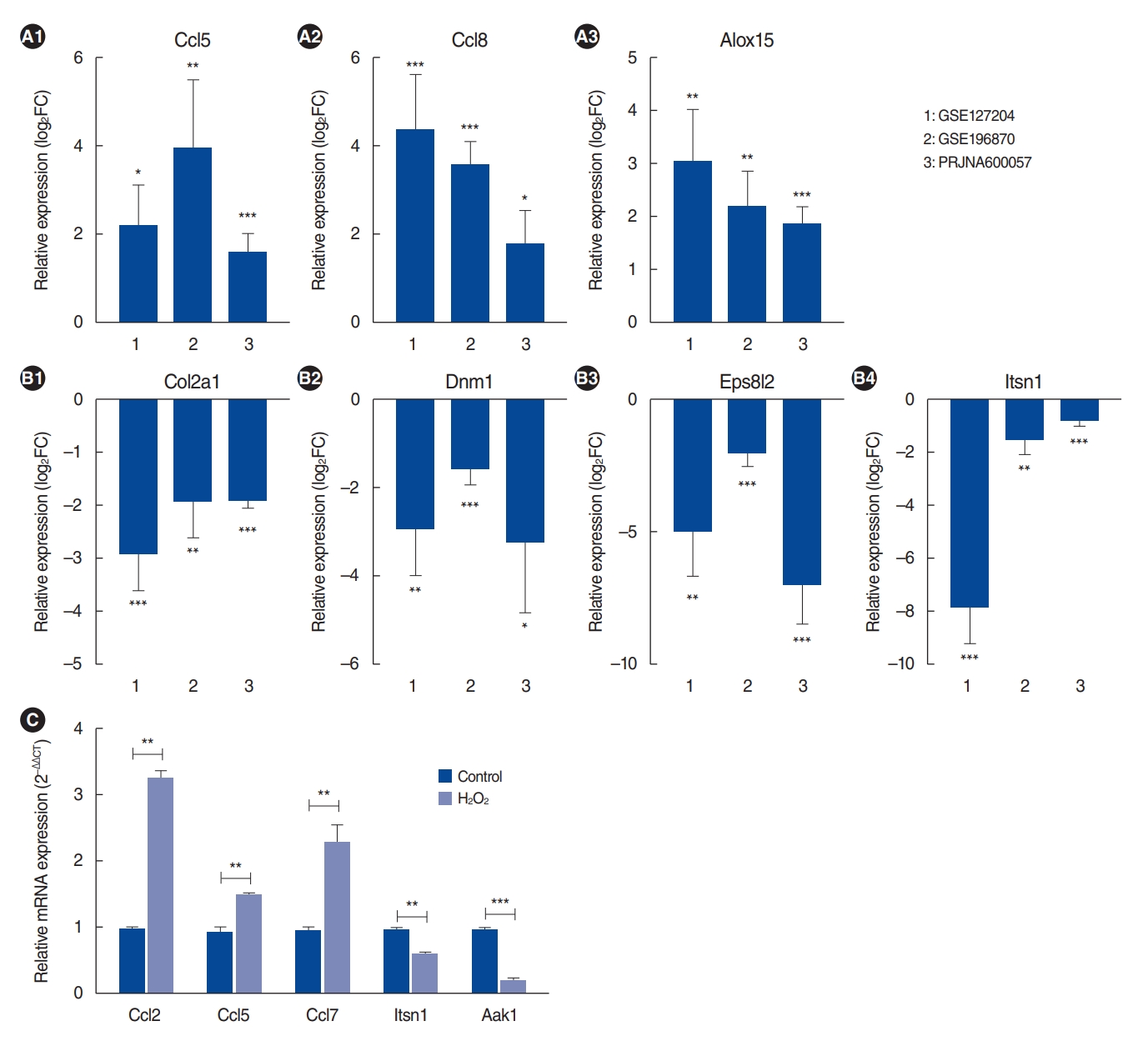
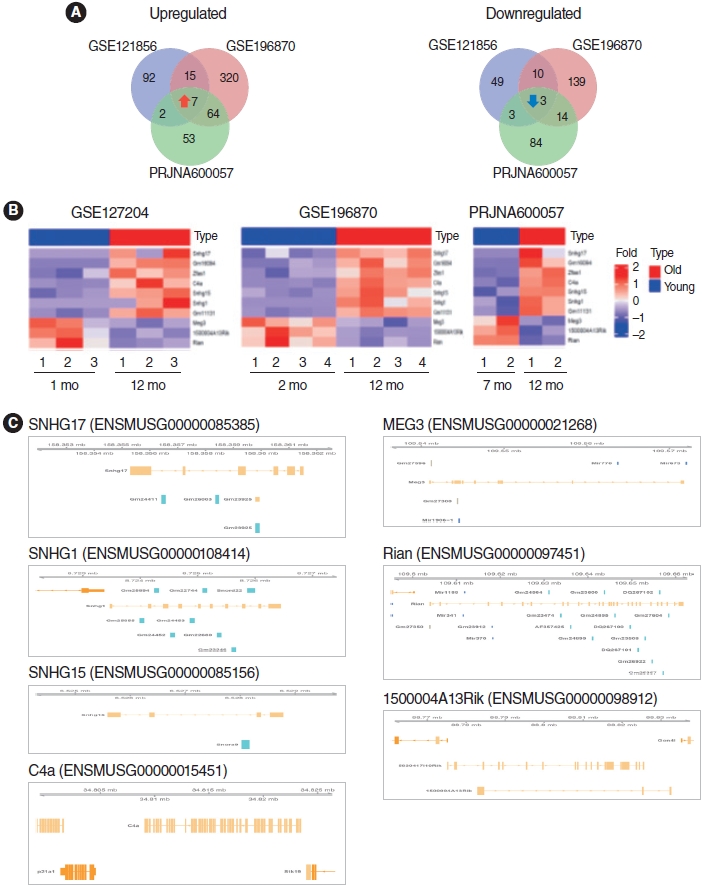
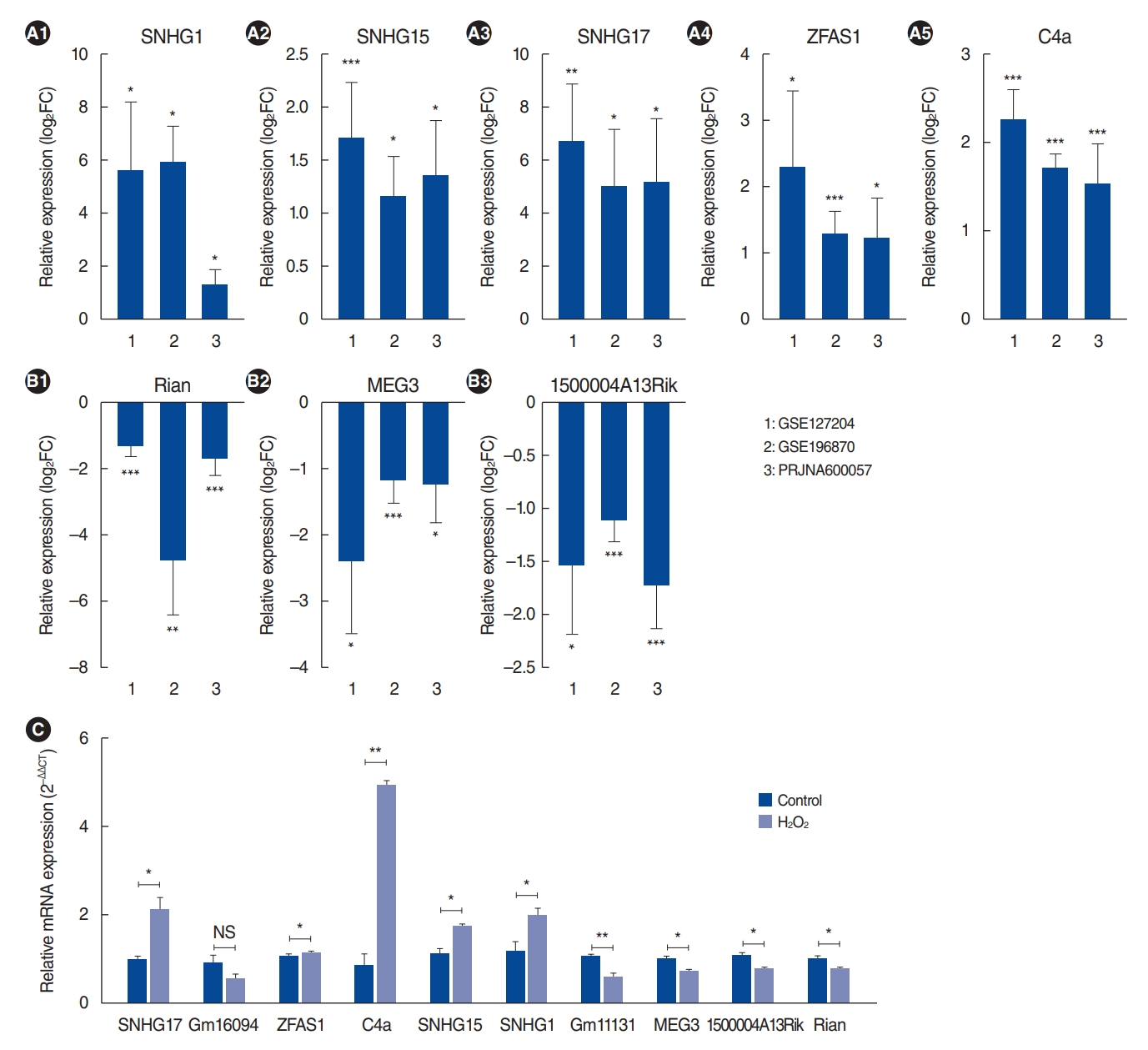
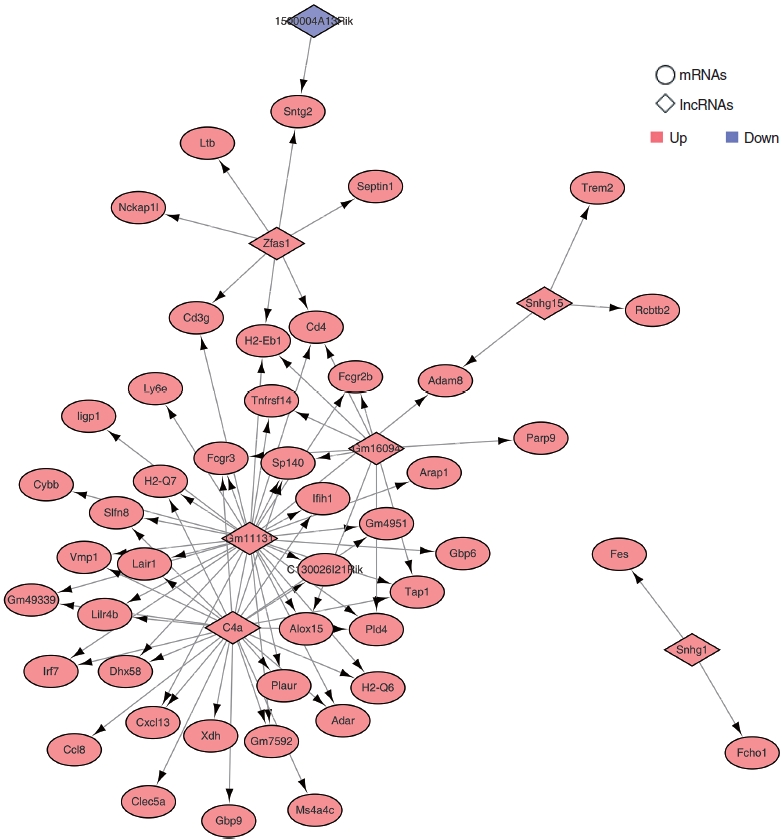
. We identified 112 common mRNAs and 10 common lncRNAs in the cochleae of aged mice compared to young mice. GO analysis showed that the 112 upregulated mRNAs were enriched in the defense response pathway. When we performed qRT-PCR with 1 mM H2O2-treated House Ear Institute-Organ of Corti 1 (HEI-OC1) cells, the qRT-PCR results were consistent with the RNA-sequencing analysis data. lncRNA-mRNA networks were constructed using the 10 common lncRNAs and 112 common mRNAs in ARHL.
Conclusion
. Our study provides a comprehensive understanding of the common mRNA and lncRNA expression profiles in ARHL. Knowledge of ARHL-associated mRNAs and lncRNAs could be useful for better understanding ARHL and these mRNAs and lncRNAs might be a potential therapeutic target for preventing ARHL.
Keyword
Figure
Reference
-
1. Gates GA, Mills JH. Presbycusis. Lancet. 2005; Sep. 366(9491):1111–20.
Article2. Keithley EM. Pathology and mechanisms of cochlear aging. J Neurosci Res. 2020; Sep. 98(9):1674–84.
Article3. Di Stazio M, Morgan A, Brumat M, Bassani S, Dell’Orco D, Marino V, et al. New age-related hearing loss candidate genes in humans: an ongoing challenge. Gene. 2020; Jun. 742:144561.
Article4. Ames BN. Mitochondrial decay, a major cause of aging, can be delayed. J Alzheimers Dis. 2004; Apr. 6(2):117–21.
Article5. Steyger PS, Cunningham LL, Esquivel CR, Watts KL, Zuo J. Editorial: cellular mechanisms of ototoxicity. Front Cell Neurosci. 2018; Mar. 12:75.
Article6. Menardo J, Tang Y, Ladrech S, Lenoir M, Casas F, Michel C, et al. Oxidative stress, inflammation, and autophagic stress as the key mechanisms of premature age-related hearing loss in SAMP8 mouse Cochlea. Antioxid Redox Signal. 2012; Feb. 16(3):263–74.
Article7. Loughrey DG, Kelly ME, Kelley GA, Brennan S, Lawlor BA. Association of age-related hearing loss with cognitive function, cognitive impairment, and dementia: a systematic review and meta-analysis. JAMA Otolaryngol Head Neck Surg. 2018; Feb. 144(2):115–26.
Article8. Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013; Jun. 54(6):301–304. 301, 303-4.
Article9. Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, et al. Long non-coding RNAs: mechanism of action and functional utility. Noncoding RNA Res. 2016; Nov. 1(1):43–50.
Article10. Sun Q, Song YJ, Prasanth KV. One locus with two roles: microRNA-independent functions of microRNA-host-gene locus-encoded long noncoding RNAs. Wiley Interdiscip Rev RNA. 2021; May. 12(3):e1625.
Article11. Li Q, Zang Y, Sun Z, Zhang W, Liu H. Long noncoding RNA Gm44593 attenuates oxidative stress from age-related hearing loss by regulating miR-29b/WNK1. Bioengineered. 2022; Jan. 13(1):573–82.
Article12. Su Z, Xiong H, Pang J, Lin H, Lai L, Zhang H, et al. LncRNA AW112010 promotes mitochondrial biogenesis and hair cell survival: implications for age-related hearing loss. Oxid Med Cell Longev. 2019; Oct. 2019:6150148.
Article13. Hao S, Wang L, Zhao K, Zhu X, Ye F. Rs1894720 polymorphism in MIAT increased susceptibility to age-related hearing loss by modulating the activation of miR-29b/SIRT1/PGC-1α signaling. J Cell Biochem. 2019; Apr. 120(4):4975–86.14. Xie W, Shu T, Peng H, Liu J, Li C, Wang M, et al. LncRNA H19 inhibits oxidative stress injury of cochlear hair cells by regulating miR-653-5p/SIRT1 axis. Acta Biochim Biophys Sin (Shanghai). 2022; Mar. 54(3):332–9.
Article15. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; Aug. 30(15):2114–20.
Article16. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; Sep. 22(9):1760–74.
Article17. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017; Apr. 14(4):417–9.
Article18. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015; Dec. 4:1521.
Article19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; Dec. 15(12):550.
Article20. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; Sep. 32(18):2847–9.
Article21. Su Z, Xiong H, Liu Y, Pang J, Lin H, Zhang W, et al. Transcriptomic analysis highlights cochlear inflammation associated with age-related hearing loss in C57BL/6 mice using next generation sequencing. PeerJ. 2020; Aug. 8:e9737.
Article22. Chen P, Hao JJ, Li MW, Bai J, Guo YT, Liu Z, et al. Integrative functional transcriptomic analyses implicate shared molecular circuits in sensorineural hearing loss. Front Cell Neurosci. 2022; Mar. 16:857344.
Article23. Lin H, Xiong H, Su Z, Pang J, Lai L, Zhang H, et al. Inhibition of DRP-1-dependent mitophagy promotes cochlea hair cell senescence and exacerbates age-related hearing loss. Front Cell Neurosci. 2019; Dec. 13:550.
Article24. Qi F, Zhang R, Chen J, Zhao F, Sun Y, Du Z, et al. Down-regulation of Cav1.3 in auditory pathway promotes age-related hearing loss by enhancing calcium-mediated oxidative stress in male mice. Aging (Albany NY). 2019; Aug. 11(16):6490–502.
Article25. Park C, Thein P, Kalinec G, Kalinec F. HEI-OC1 cells as a model for investigating prestin function. Hear Res. 2016; May. 335:9–17.
Article26. Kalinec GM, Park C, Thein P, Kalinec F. Working with auditory HEIOC1 cells. J Vis Exp. 2016; Sep. (115):54425.
Article27. Zhang Q, Liu H, McGee J, Walsh EJ, Soukup GA, He DZ. Identifying microRNAs involved in degeneration of the organ of corti during age-related hearing loss. PLoS One. 2013; Apr. 8(4):e62786.
Article28. Orrenius S. Reactive oxygen species in mitochondria-mediated cell death. Drug Metab Rev. 2007; 39(2-3):443–55.
Article29. Yu J, Wang Y, Liu P, Li Q, Sun Y, Kong W. Mitochondrial DNA common deletion increases susceptibility to noise-induced hearing loss in a mimetic aging rat model. Biochem Biophys Res Commun. 2014; Oct. 453(3):515–20.
Article30. Zimta AA, Tigu AB, Braicu C, Stefan C, Ionescu C, Berindan-Neagoe I. An emerging class of long non-coding rna with oncogenic role arises from the snorna host genes. Front Oncol. 2020; Apr. 10:389.
Article31. Ge BH, Li GC. Long non-coding RNA SNHG17 promotes proliferation, migration and invasion of glioma cells by regulating the miR23b-3p/ZHX1 axis. J Gene Med. 2020; Nov. 22(11):e3247.
Article32. Ji YY, Meng M, Miao Y. lncRNA SNHG1 promotes progression of cervical cancer through miR-195/NEK2 axis. Cancer Manag Res. 2020; Nov. 12:11423–33.33. Chen Y, Sheng HG, Deng FM, Cai LL. Downregulation of the long noncoding RNA SNHG1 inhibits tumor cell migration and invasion by sponging miR-195 through targeting Cdc42 in oesophageal cancer. Kaohsiung J Med Sci. 2021; Mar. 37(3):181–91.34. Tong J, Ma X, Yu H, Yang J. SNHG15: a promising cancer-related long noncoding RNA. Cancer Manag Res. 2019; Jul. 11:5961–9.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Rehabilitation of Age-Related Hearing Loss Patients
- The Expression of AGO2 and DGCR8 in Idiopathic Sudden Sensorineural Hearing Loss
- Newborn Hearing Loss and Newborn Hearing Screening
- Dicer Is Down-regulated and Correlated with Drosha in Idiopathic Sudden Sensorineural Hearing Loss
- Revisiting Age-Related Normative Hearing Levels in Korea