Korean J Physiol Pharmacol.
2023 May;27(3):197-208. 10.4196/kjpp.2023.27.3.197.
LINC00562 drives gastric cancer development by regulating miR-4636–AP1S3 axis
- Affiliations
-
- 1Digestion Medicine Department, Wuchang Hospital Affiliated to Wuhan University of Science and Technology, Wuhan 430063, China
- KMID: 2541632
- DOI: http://doi.org/10.4196/kjpp.2023.27.3.197
Abstract
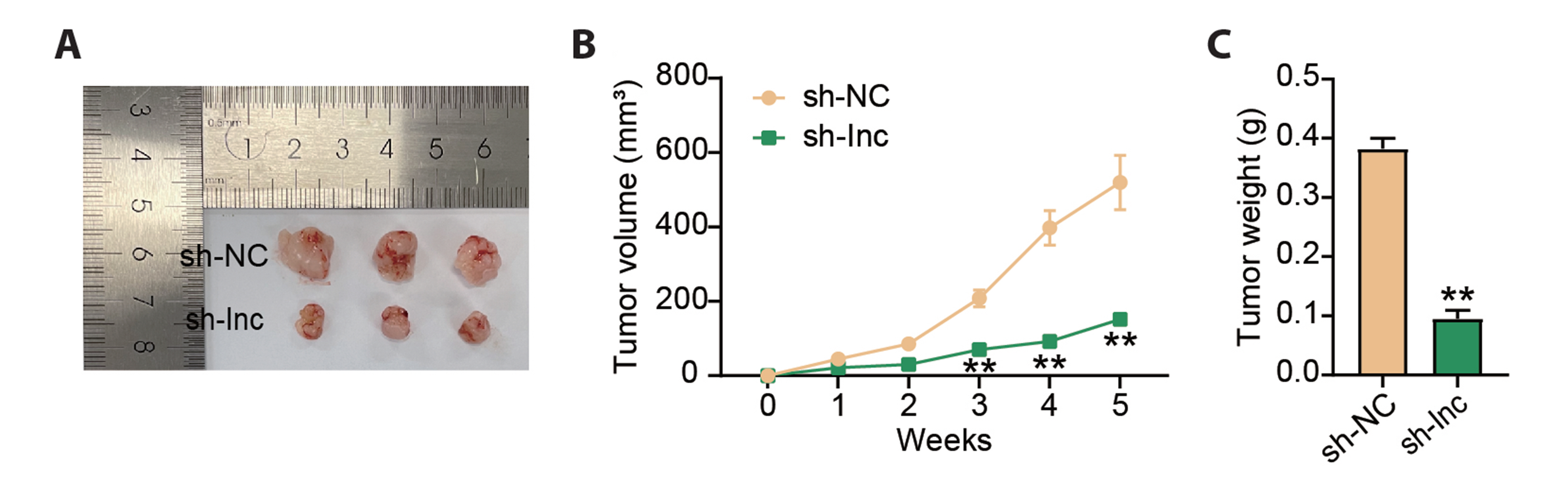
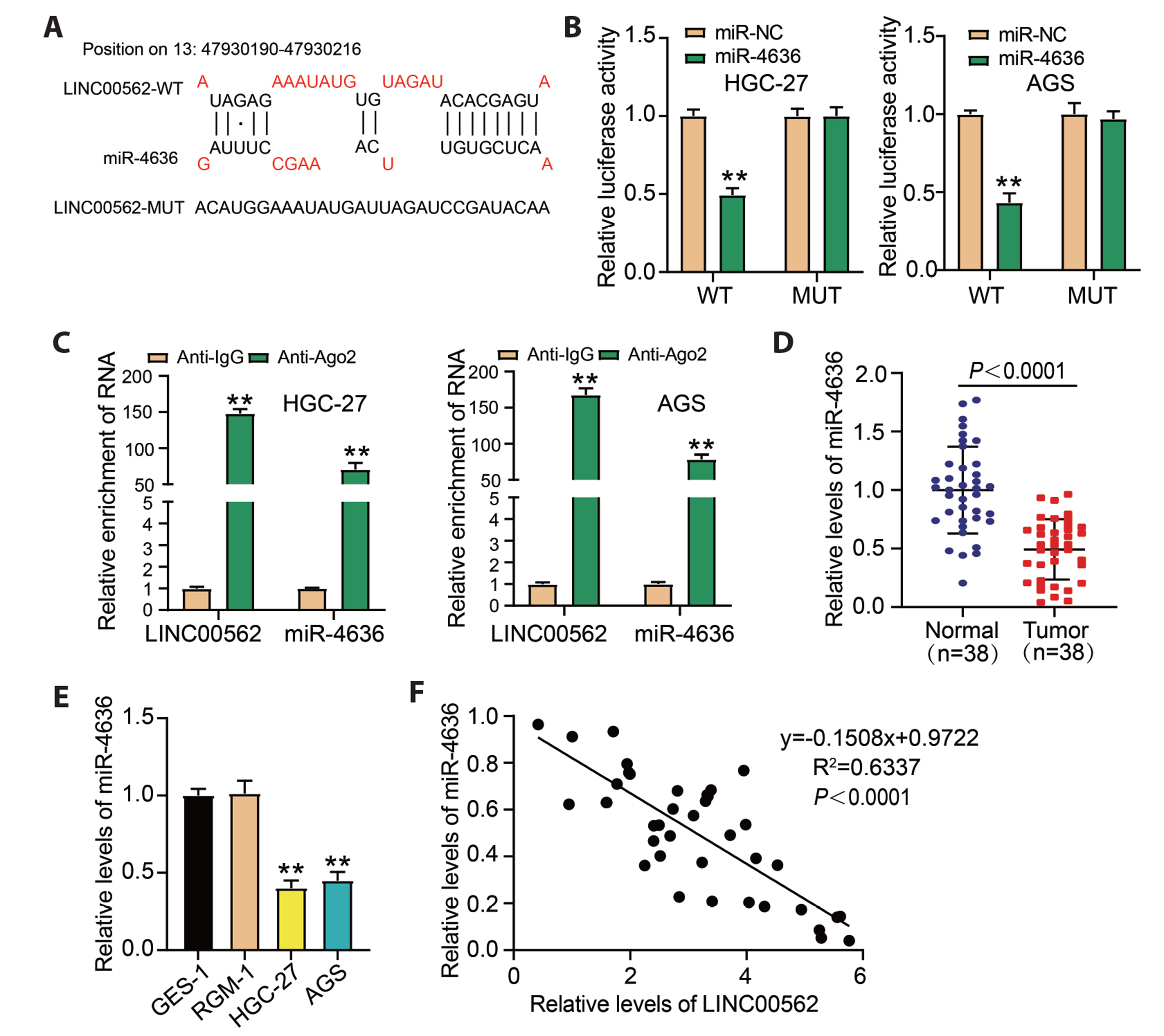
- Dysregulation of certain long non-coding RNAs may facilitate tumor initiation and progression. However, numerous carcinogenesis-related long non-coding RNAs have not been characterized. The goal of this study was to elucidate the role of LINC00562 in gastric cancer (GC). The expression of LINC00562 was analyzed using real-time quantitative PCR and Western blotting. The proliferative capacity of GC cells was determined using Cell Counting Kit-8 and colony-formation assays. The migration of GC cells were evaluated using wound-healing assays. The apoptosis of GC cells was assessed by measuring the expression levels of apoptosis-related proteins (Bax and Bcl-2). Xenograft models in nude mice were constructed for in vivo functional analysis of LINC00562. The binding relationship between miR-4636 and LINC00562 or adaptor protein complex 1 sigma 3 (AP1S3), obtained from public databases, was confirmed using dual-luciferase and RNA-binding protein immunoprecipitation experiments. LINC00562 was expressed in GC cells at high levels. Knockdown of LINC00562 repressed GC cell growth and migration, promoted apoptosis in vitro, and inhibited tumor growth in nude mouse models. LINC00562 directly targeted miR-4636, and miR-4636 depletion restored the GC cell behavior inhibited by LINC00562 absence. AP1S3, an oncogene, binds to miR-4636. MiR-4636 downregulation increased AP1S3 level, restoring GC cell malignant behaviors inhibited by AP1S3 downregulation. Thus, LINC00562 exerts carcinogenic effects on GC development by targeting miR-4636-mediated AP1S3 signaling.
Figure
Reference
-
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021; Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. DOI: 10.3322/caac.21660. PMID: 33538338.
Article2. Lim B, Kim JH, Kim M, Kim SY. 2016; Genomic and epigenomic heterogeneity in molecular subtypes of gastric cancer. World J Gastroenterol. 22:1190–1201. DOI: 10.3748/wjg.v22.i3.1190. PMID: 26811657. PMCID: PMC4716030.
Article3. Ikeguchi M, Miyatani K, Takaya S, Matsunaga T, Fukumoto Y, Osaki T, Saito H, Wakatsuki T. 2016; Role of surgery in the management for gastric cancer with synchronous distant metastases. Indian J Surg Oncol. 7:32–36. DOI: 10.1007/s13193-015-0428-6. PMID: 27065679. PMCID: PMC4811814.
Article4. Canale M, Casadei-Gardini A, Ulivi P, Arechederra M, Berasain C, Lollini PL, Fernández-Barrena MG, Avila MA. 2020; Epigenetic mechanisms in gastric cancer: potential new therapeutic opportunities. Int J Mol Sci. 21:5500. DOI: 10.3390/ijms21155500. PMID: 32752096. PMCID: PMC7432799. PMID: 80935d64283843258726a4a6040830b3.
Article5. Ma J, Hong L, Chen Z, Nie Y, Fan D. 2014; Epigenetic regulation of microRNAs in gastric cancer. Dig Dis Sci. 59:716–723. DOI: 10.1007/s10620-013-2939-8. PMID: 24248419.
Article6. Cao J. 2014; The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 16:11. DOI: 10.1186/1480-9222-16-11. PMID: 25276098. PMCID: PMC4177375.
Article7. Li M, Liu Y, Zhang X, Liu J, Wang P. 2018; Transcriptomic analysis of high-throughput sequencing about circRNA, lncRNA and mRNA in bladder cancer. Gene. 677:189–197. DOI: 10.1016/j.gene.2018.07.041. PMID: 30025927.
Article8. Bhan A, Soleimani M, Mandal SS. 2017; Long noncoding RNA and cancer: a new paradigm. Cancer Res. 77:3965–3981. DOI: 10.1158/0008-5472.CAN-16-2634. PMID: 28701486. PMCID: PMC8330958.
Article9. Wang M, Gu J, Zhang X, Yang J, Zhang X, Fang X. 2021; Long non-coding RNA DANCR in cancer: roles, mechanisms, and implications. Front Cell Dev Biol. 9:753706. DOI: 10.3389/fcell.2021.753706. PMID: 34722539. PMCID: PMC8554091. PMID: 2ee9f53482274f1ab864ccbd6e1e7a15.
Article10. Tian T, Wang M, Lin S, Guo Y, Dai Z, Liu K, Yang P, Dai C, Zhu Y, Zheng Y, Xu P, Zhu W, Dai Z. 2018; The impact of lncRNA dysregulation on clinicopathology and survival of breast cancer: a systematic review and meta-analysis. Mol Ther Nucleic Acids. 12:359–369. DOI: 10.1016/j.omtn.2018.05.018. PMID: 30195774. PMCID: PMC6037885.
Article11. Fattahi S, Kosari-Monfared M, Golpour M, Emami Z, Ghasemiyan M, Nouri M, Akhavan-Niaki H. 2020; LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: a novel approach to personalized medicine. J Cell Physiol. 235:3189–3206. DOI: 10.1002/jcp.29260. PMID: 31595495.
Article12. López-Urrutia E, Bustamante Montes LP, Ladrón de Guevara Cervantes D, Pérez-Plasencia C, Campos-Parra AD. 2019; Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: deciphering molecular mechanisms of master regulators in cancer. Front Oncol. 9:669. DOI: 10.3389/fonc.2019.00669. PMID: 31404273. PMCID: PMC6670781. PMID: 6987929dd3cf42c2afb8317153f61b43.
Article13. Karagkouni D, Paraskevopoulou MD, Tastsoglou S, Skoufos G, Karavangeli A, Pierros V, Zacharopoulou E, Hatzigeorgiou AG. 2020; DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 48:D101–D110. DOI: 10.1093/nar/gkz1036. PMID: 31732741. PMCID: PMC7145509.
Article14. Agarwal V, Bell GW, Nam JW, Bartel DP. 2015; Predicting effective microRNA target sites in mammalian mRNAs. Elife. 4:e05005. DOI: 10.7554/eLife.05005. PMID: 26267216. PMCID: PMC4532895. PMID: a795d72f226e46a8803f727538963f18.
Article15. Xiao S, Guo J, Zhang W, Hu X, Wang R, Chen Z, Lai C. 2022; A six-microRNA signature nomogram for preoperative prediction of tumor deposits in colorectal cancer. Int J Gen Med. 15:675–687. DOI: 10.2147/IJGM.S346790. PMID: 35082517. PMCID: PMC8785134.
Article16. Tang J, Hu Y, Zhang C, Gong C. 2021; miR-4636 inhibits tumor cell proliferation, migration and invasion, and serves as a candidate clinical biomarker for gastric cancer. Oncol Lett. 21:33. DOI: 10.3892/ol.2020.12294. PMID: 33262825. PMCID: PMC7693299.
Article17. Yin S, Yang M, Li X, Zhang K, Tian J, Luo C, Bai R, Lu Y, Wang M. 2020; Peripheral blood circulating microRNA-4636/-143 for the prognosis of cervical cancer. J Cell Biochem. 121:596–608. DOI: 10.1002/jcb.29305. PMID: 31407404.
Article18. Duncan MC. 2022; New directions for the clathrin adaptor AP-1 in cell biology and human disease. Curr Opin Cell Biol. 76:102079. DOI: 10.1016/j.ceb.2022.102079. PMID: 35429729.
Article19. Toda H, Kurozumi S, Kijima Y, Idichi T, Shinden Y, Yamada Y, Arai T, Maemura K, Fujii T, Horiguchi J, Natsugoe S, Seki N. 2018; Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: antitumor miR-204-5p targets AP1S3. J Hum Genet. 63:1197–1210. DOI: 10.1038/s10038-018-0510-3. PMID: 30228364.
Article20. Ye T, Cheng Y, Li C. 2021; Adaptor protein complex 1 sigma 3 is highly expressed in glioma and could enhance its progression. Comput Math Methods Med. 2021:5086236. DOI: 10.1155/2021/5086236. PMID: 34367317. PMCID: PMC8346305.
Article21. Livak KJ, Schmittgen TD. 2001; Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25:402–408. DOI: 10.1006/meth.2001.1262. PMID: 11846609.
Article22. Chen Q, Yao YT, Xu H, Chen YB, Gu M, Cai ZK, Wang Z. 2016; SPOCK1 promotes tumor growth and metastasis in human prostate cancer. Drug Des Devel Ther. 10:2311–2321. DOI: 10.2147/DDDT.S91321. PMID: 27486308. PMCID: PMC4958368.23. Zhou L, Zhu Y, Sun D, Zhang Q. 2020; Emerging roles of long non-coding RNAs in the tumor microenvironment. Int J Biol Sci. 16:2094–2103. DOI: 10.7150/ijbs.44420. PMID: 32549757. PMCID: PMC7294937.
Article24. Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, Rao X, Li M, Sun M, Jiang M, Xu Y, Teng L, Jin Y, Si J, Liu W, Kang Y, Zhou T. 2019; Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 156:676–691.e11. DOI: 10.1053/j.gastro.2018.10.054. PMID: 30445010.
Article25. Yu Y, Li L, Zheng Z, Chen S, Chen E, Hu Y. 2017; Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. J Cell Mol Med. 21:955–967. DOI: 10.1111/jcmm.13035. PMID: 27878953. PMCID: PMC5387161.
Article26. Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, Zhao G, Zhang ZZ. 2019; The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 18:115. Erratum in: Mol Cancer. 2021;20:120. DOI: 10.1186/s12943-019-1032-0. PMID: 31272462. PMCID: PMC6609402. PMID: ee6fb104778948bc9014fe573397dfe8.
Article27. Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, Okato A, Arai T, Kita Y, Mataki Y, Kijima Y, Maemura K, Natsugoe S. 2018; Molecular pathogenesis of pancreatic ductal adenocarcinoma: impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 109:2013–2026. DOI: 10.1111/cas.13610. PMID: 29660218. PMCID: PMC5989856.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Combined Detection of Serum MiR-221-3p and MiR-122-5p Expression in Diagnosis and Prognosis of Gastric Cancer
- LINC00703 Acts as a Tumor Suppressor via Regulating miR-181a/KLF6 Axis in Gastric Cancer
- RETRACTION: Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN
- Circ_0081143 Contributes to Gastric Cancer Malignant Development and Doxorubicin Resistance by Elevating the Expression of YES1 by Targeting mziR-129-2-3p
- Retraction: Circ_0075960 targets the miR-202-5p/CTNND1 axis to promote the growth and migration of endometrial carcinoma cells via regulating Wnt/β-catenin signaling activity