Lab Med Online.
2022 Jan;12(1):11-19. 10.47429/lmo.2022.12.1.11.
Comparison of Automated and Manual Gating of Lymphocyte Subsets in Hematopoietic Stem Cell Transplantation Recipients
- Affiliations
-
- 1Department of Laboratory Medicine, Catholic Kwandong University International St. Mary’s Hospital, Incheon, Korea
- 2Department of Laboratory Medicine, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2538578
- DOI: http://doi.org/10.47429/lmo.2022.12.1.11
Abstract
- Background
Lymphocyte subset analysis is essential to evaluate the engraftment status in hematopoietic stem cell transplantation (HSCT). Automated gating tools are widely used for flow cytometry analysis. Unlike healthy individuals, different cell populations and aberrant expressions may occur in HSCT samples. In the present study, we evaluated the applicability of automated gating in HSCT recipients by comparing it to expert-based manual gating.
Methods
Lymphocyte subset was performed using Beckman Coulter Navios (Beckman Coulter, USA) flow cytometry. Data files from 22 patients with hematologic malignancies were analyzed in parallel by manual gating and automated gating using Navios Tetra software. Quality control results and reproducibility were evaluated using IMMUNO-TROL controls.
Results
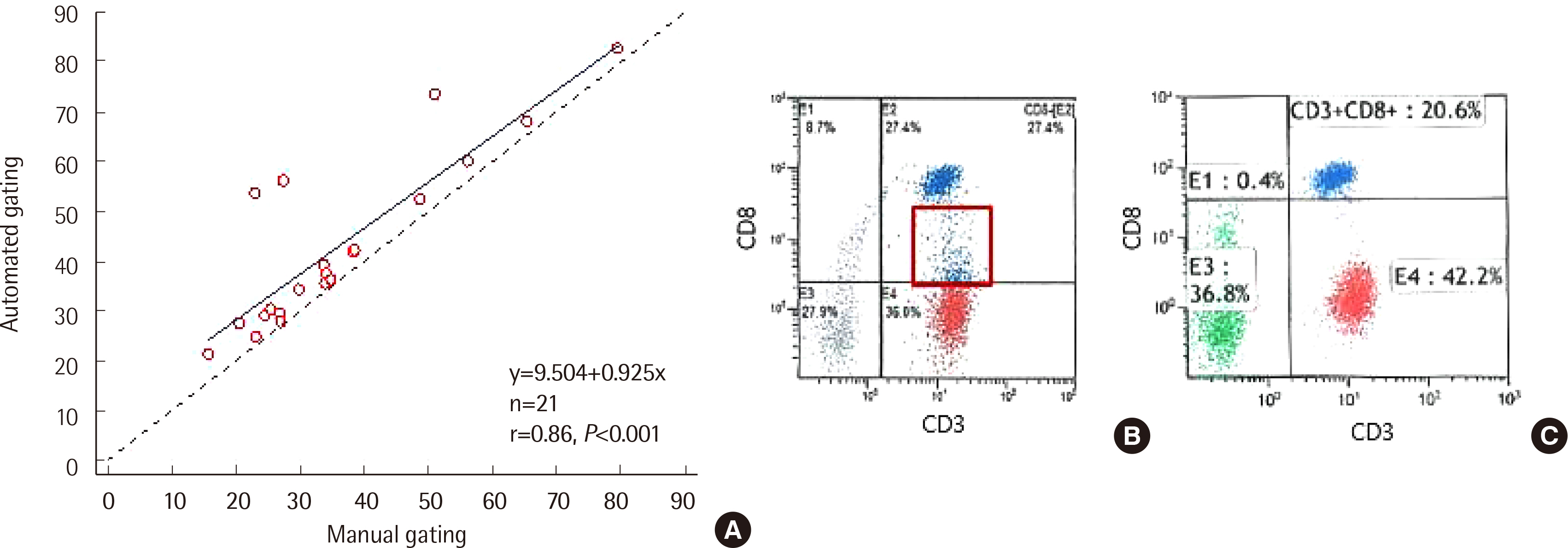
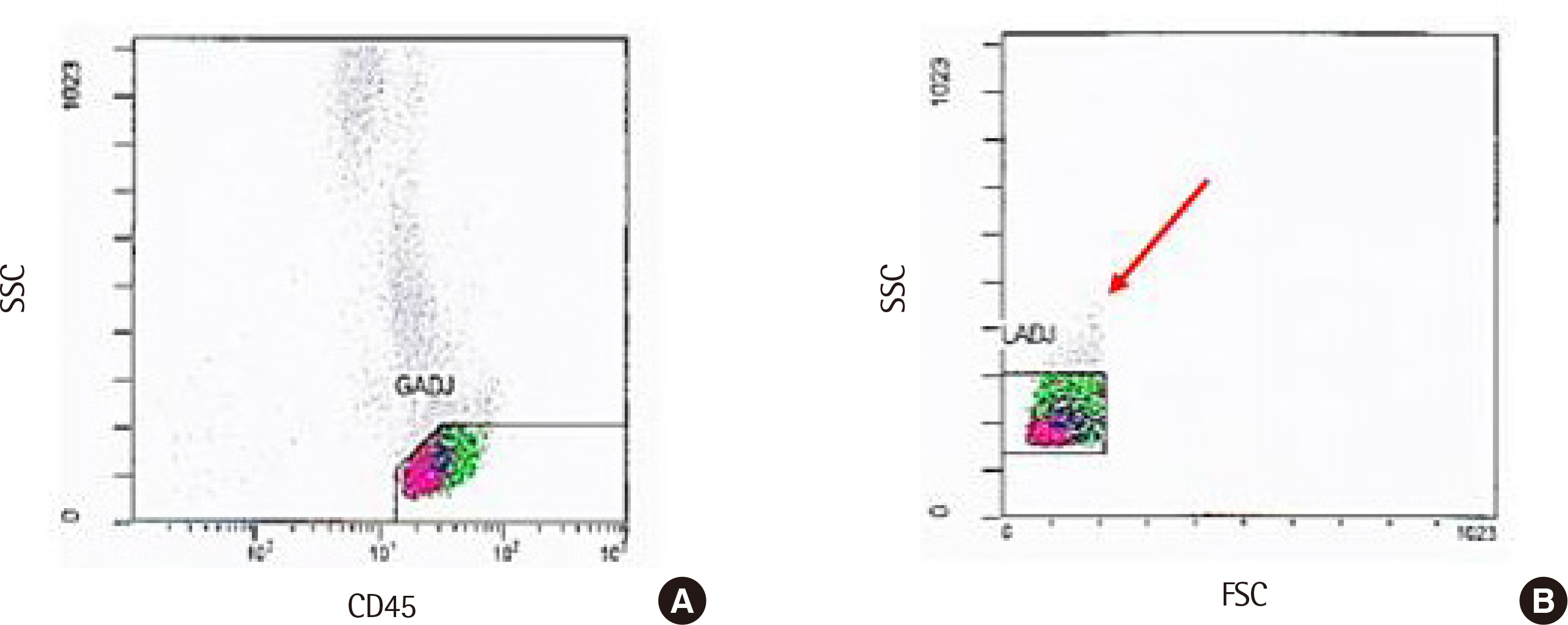
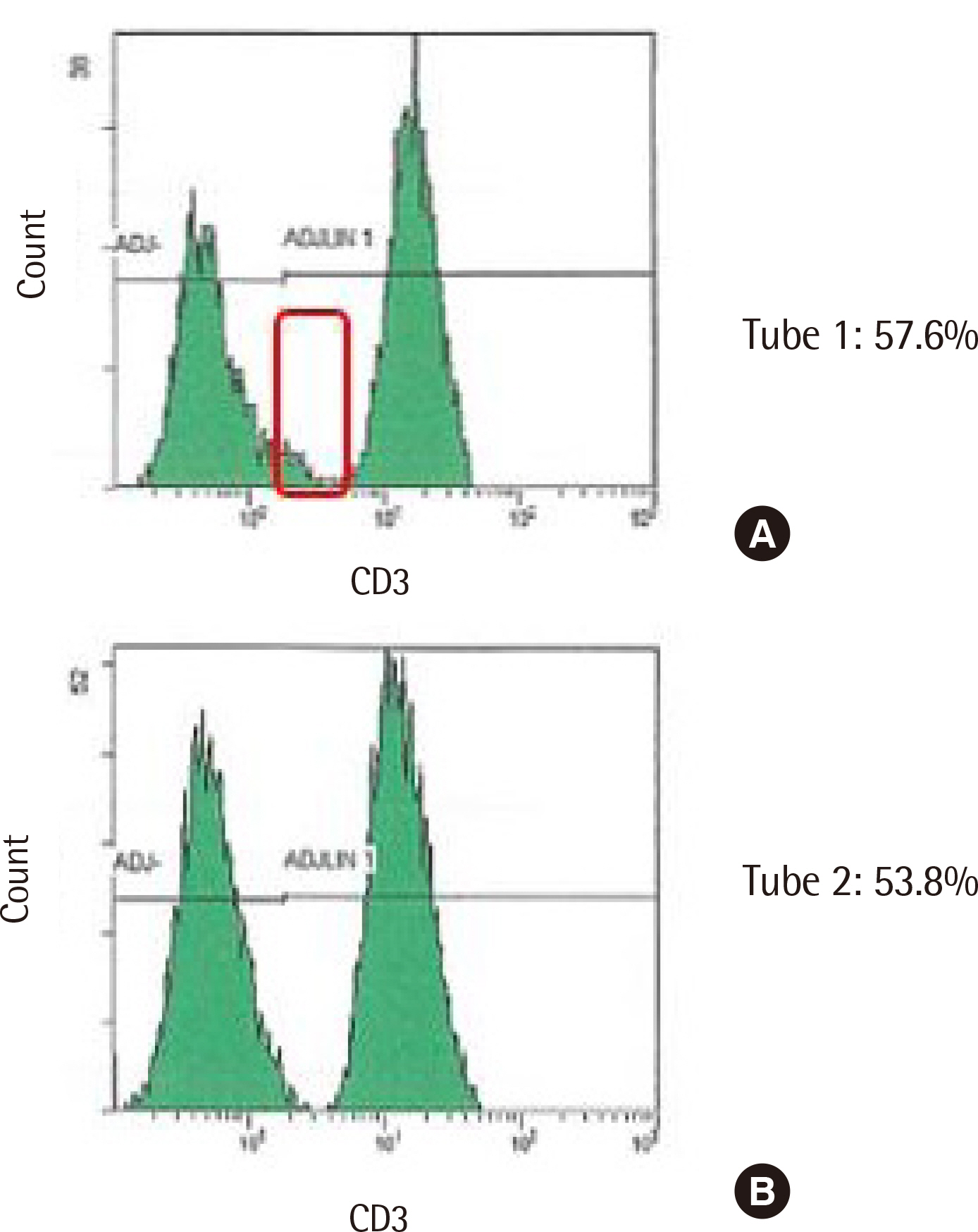
Spearman rank correlation coefficients between the two gating methods were >0.970 in all cell populations except CD8+ T cells. CD8+ T cell counts via automated gating were higher than those of manual gating in all cases due to the T cell populations with reduced CD8 expression. Automated gating program failed to identify CD4+CD8+ double-positive T cell population. Moreover, it excluded certain lymphocytes with low forward scatter (FSC) and high side scatter (SSC). Furthermore, two HSCT recipients revealed a high percentage of CD56−CD16+ NK cells, we found the need to add CD16 reagent to the Navios system. All coefficients of variation were <10% except for CD56+ NK cells via automated gating.
Conclusions
Manual gating confirmation via flow cytometry histogram is necessary to identify the aberrant phenotypes and unexpected cell populations in HSCT recipients.
Keyword
Figure
Reference
-
1. Peggs KS, Mackinnon S. 2004; Immune reconstitution following haematopoietic stem cell transplantation. Br J Haematol. 124:407–20. DOI: 10.1046/j.1365-2141.2003.04767.x. PMID: 14984491.
Article2. Storek J, Dawson MA, Storer B, Stevens-Ayers T, Maloney DG, Marr KA, et al. 2001; Immune reconstitution after allogeneic marrow transplantation compared with blood stem cell transplantation. Blood. 97:3380–9. DOI: 10.1182/blood.V97.11.3380. PMID: 11369627.
Article3. Savani BN, Rezvani K, Mielke S, Montero A, Kurlander R, Carter CS, et al. 2006; Factors associated with early molecular remission after T cell-depleted allogeneic stem cell transplantation for chronic myelogenous leukemia. Blood. 107:1688–95. DOI: 10.1182/blood-2005-05-1897. PMID: 16131570. PMCID: PMC1895415.
Article4. Kim DH, Sohn SK, Won DI, Lee NY, Suh JS, Lee KB. 2006; Rapid helper T-cell recovery above 200×106/l at 3 months correlates to successful transplant outcomes after allogeneic stem cell transplantation. Bone Marrow Transplant. 37:1119–28. DOI: 10.1038/sj.bmt.1705381. PMID: 16699530.5. Williams KM, Gress RE. 2008; Immune reconstitution and implications for immunotherapy following haematopoietic stem cell transplantation. Best Pract Res Clin Haematol. 21:579–96. DOI: 10.1016/j.beha.2008.06.003. PMID: 18790456. PMCID: PMC2577193.
Article6. Brinkman RR. 2020; Improving the rigor and reproducibility of flow cytometry-based clinical research and trials through automated data analysis. Cytometry A. 97:107–12. DOI: 10.1002/cyto.a.23883. PMID: 31515945. PMCID: PMC8043840.
Article7. Montante S, Brinkman RR. 2019; Flow cytometry data analysis: Recent tools and algorithms. Int J Lab Hematol. 41(Suppl 1):56–62. DOI: 10.1111/ijlh.13016. PMID: 31069980.
Article8. Frelinger J, Ottinger J, Gouttefangeas C, Chan C. 2010; Modeling flow cytometry data for cancer vaccine immune monitoring. Cancer Immunol Immunother. 59:1435–41. DOI: 10.1007/s00262-010-0883-4. PMID: 20563720. PMCID: PMC2892609.
Article9. Meehan S, Walther G, Moore W, Orlova D, Meehan C, Parks D, et al. 2014; AutoGate: automating analysis of flow cytometry data. Immunol Res. 58:218–23. DOI: 10.1007/s12026-014-8519-y. PMID: 24825775. PMCID: PMC4464812.
Article10. Flores-Montero J, Grigore G, Fluxá R, Hernández J, Fernandez P, Almeida J, et al. 2019; EuroFlow Lymphoid Screening Tube (LST) data base for automated identification of blood lymphocyte subsets. J Immunol Methods. 475:112662. DOI: 10.1016/j.jim.2019.112662. PMID: 31454495.
Article11. Verschoor CP, Lelic A, Bramson JL, Bowdish DM. 2015; An introduction to automated flow cytometry gating tools and their implementation. Front Immunol. 6:380. DOI: 10.3389/fimmu.2015.00380. PMID: 26284066. PMCID: PMC4515551.
Article12. Mair F, Hartmann FJ, Mrdjen D, Tosevski V, Krieg C, Becher B. 2016; The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol. 46:34–43. DOI: 10.1002/eji.201545774. PMID: 26548301.
Article13. Conrad VK, Dubay CJ, Malek M, Brinkman RR, Koguchi Y, Redmond WL. 2019; Implementation and validation of an automated flow cytometry analysis pipeline for human immune profiling. Cytometry A. 95:183–91. DOI: 10.1002/cyto.a.23664. PMID: 30570217.
Article14. Aghaeepour N, Finak G, Hoos H, Mosmann TR, Brinkman R, Gottardo R, et al. 2013; Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 10:228–38. DOI: 10.1038/nmeth.2365. PMID: 23396282. PMCID: PMC3906045.
Article15. Mandy FF, Nicholson JK, McDougal JS. 2003; Guidelines for performing single-platform absolute CD4+ T-cell determinations with CD45 gating for persons infected with human immunodeficiency virus. Centers for Disease Control and Prevention. MMWR Recomm Rep. 52:1–13. PMID: 12583540.16. Linskens E, Diks AM, Neirinck J, Perez-Andres M, De Maertelaere E, Berkowska MA, et al. 2020; Improved standardization of flow cytometry diagnostic screening of primary immunodeficiency by software-based automated gating. Front Immunol. 11:584646. DOI: 10.3389/fimmu.2020.584646. PMID: 33224147. PMCID: PMC7667243.
Article17. Trautmann A, Rückert B, Schmid-Grendelmeier P, Niederer E, Bröcker EB, Blaser K, et al. 2003; Human CD8 T cells of the peripheral blood contain a low CD8 expressing cytotoxic/effector subpopulation. Immunology. 108:305–12. DOI: 10.1046/j.1365-2567.2003.01590.x. PMID: 12603596. PMCID: PMC1782903.
Article18. Ouyang L, Li X, Liang Z, Yang D, Gong F, Shen G, et al. 2013; CD8low T-cell subpopulation is increased in patients with chronic hepatitis B virus infection. Mol Immunol. 56:698–704. DOI: 10.1016/j.molimm.2013.07.003. PMID: 23933510.19. Marins-Dos-Santos A, Olivieri BP, Ferreira-Reis R, de Meis J, Silva AA, de Araújo-Jorge TC, et al. 2020; CD8low T cells expanded following acute Trypanosoma cruzi infection and benznidazole treatment are a relevant subset of IFN-gamma producers. PLoS Negl Trop Dis. 14:e0008969. DOI: 10.1371/journal.pntd.0008969. PMID: 33347472. PMCID: PMC7785226.20. Liu B, Ma Y, Zhang Y, Zhang C, Yi J, Zhuang R, et al. 2015; CD8low CD100- T cells identify a novel CD8 T cell subset associated with viral control during human Hantaan virus infection. J Virol. 89:11834–44. DOI: 10.1128/JVI.01610-15. PMID: 26378166. PMCID: PMC4645342.21. Xu H, Wang X, Lackner AA, Veazey RS. 2013; CD8 down-regulation and functional impairment of SIV-specific cytotoxic T lymphocytes in lymphoid and mucosal tissues during SIV infection. J Leukoc Biol. 93:943–50. DOI: 10.1189/jlb.1112580. PMID: 23519937. PMCID: PMC3656336.
Article22. Favre D, Stoddart CA, Emu B, Hoh R, Martin JN, Hecht FM, et al. 2011; HIV disease progression correlates with the generation of dysfunctional naive CD8low T cells. Blood. 117:2189–99. DOI: 10.1182/blood-2010-06-288035. PMID: 21200021. PMCID: PMC3062328.23. Overgaard NH, Jung JW, Steptoe RJ, Wells JW. 2015; CD4+/CD8+ double-positive T cells: more than just a developmental stage? J Leukoc Biol. 97:31–8. DOI: 10.1189/jlb.1RU0814-382. PMID: 25360000.24. Alhaj Hussen K, Michonneau D, Biajoux V, Keita S, Dubouchet L, Nelson E, et al. 2020; CD4+CD8+ T-lymphocytes in xenogeneic and human graft-versus-host disease. Front Immunol. 11:579776. DOI: 10.3389/fimmu.2020.579776. PMID: 33329550. PMCID: PMC7732609.25. Forconi CS, Oduor CI, Oluoch PO, Ong'echa JM, Münz C, Bailey JA, et al. 2020; A New Hope for CD56negCD16pos NK cells as unconventional cytotoxic mediators: An adaptation to chronic diseases. Front Cell Infect Microbiol. 10:162. DOI: 10.3389/fcimb.2020.00162. PMID: 32373555. PMCID: PMC7186373.
Article26. Hu PF, Hultin LE, Hultin P, Hausner MA, Hirji K, Jewett A, et al. 1995; Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity. J Acquir Immune Defic Syndr Hum Retrovirol. 10:331–40. DOI: 10.1097/00042560-199511000-00005. PMID: 7552495.27. Jacobson A, Bell F, Lejarcegui N, Mitchell C, Frenkel L, Horton H. 2013; Healthy neonates possess a CD56-negative NK cell population with reduced anti-viral activity. PLoS One. 8:e67700. DOI: 10.1371/journal.pone.0067700. PMID: 23805324. PMCID: PMC3689709.
Article28. De Angelis C, Mancusi A, Ruggeri L, Capanni M, Urbani E, Velardi A, et al. 2011; Expansion of CD56-negative, CD16-positive, KIR-expressing natural killer cells after T cell-depleted haploidentical hematopoietic stem cell transplantation. Acta Haematol. 126:13–20. DOI: 10.1159/000323661. PMID: 21411985.
Article29. Milush JM, López-Vergès S, York VA, Deeks SG, Martin JN, Hecht FM, et al. 2013; CD56negCD16+ NK cells are activated mature NK cells with impaired effector function during HIV-1 infection. Retrovirology. 10:158. DOI: 10.1186/1742-4690-10-158. PMID: 24351015. PMCID: PMC3892122.
Article30. Van Acker HH, Capsomidis A, Smits EL, Van Tendeloo VF. 2017; CD56 in the immune system: more than a marker for cytotoxicity? Front Immunol. 8:892. DOI: 10.3389/fimmu.2017.00892. PMID: 28791027. PMCID: PMC5522883.
Article31. Lu X, Kondo Y, Takamatsu H, Ohata K, Yamazaki H, Takami A, et al. 2008; CD16+ CD56- NK cells in the peripheral blood of cord blood transplant recipients: a unique subset of NK cells possibly associated with graft-versus-leukemia effect. Eur J Haematol. 81:18–25. DOI: 10.1111/j.1600-0609.2008.01073.x. PMID: 18363874.32. Clinical, Laboratory Standards Institute. 2007. Enumeration of immunologically defined cell populations by flow cytometry; Approved guideline-Second edition. CLSI document H42-A2. Clinical and Laboratory Standards Institute,;Wayne, PA: p. 18p. 21–22.33. Diallo TO, Bergeron M, Seely P, Yang X, Ding T, Plews M, et al. 2017; Automation for clinical CD4 T-cell enumeration, a desirable tool in the hands of skilled operators. Cytometry B Clin Cytom. 92:445–50. DOI: 10.1002/cyto.b.21370. PMID: 26990810.
Article34. Wood B, Jevremovic D, Bene MC, Yan M, Jacobs P, Litwin V, et al. 2013; Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS - part V - assay performance criteria. Cytometry B Clin Cytom. 84:315–23. DOI: 10.1002/cyto.b.21108. PMID: 24022854.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of Quality of Life of Autologous and Allogeneic Hematopoietic Stem Cell Transplantation Recipients
- Factors influencing lymphocyte reconstitution after allogeneic hematopoietic stem cell transplantation in children
- CD45 is Essential for Lymphocyte Gating in a T-lymphocyte Subset Assay of Bronchoalveolar Lavage Fluid by Flow Cytometry
- Opening the era of in vivo xenotransplantation model for hematopoietic stem cell transplantation
- Lineage-specific chimerism analysis in nucleated cells, T cells and natural killer cells after myeloablative allogeneic hematopoietic stem cell transplantation