Cardiovasc Prev Pharmacother.
2020 Jan;2(1):24-30. 10.36011/cpp.2020.2.e3.
Basic Concepts of a Mendelian Randomization Approach
- Affiliations
-
- 1Department of Biostatistics, Yonsei University Wonju College of Medicine, Wonju, Korea
- 2Department of Precision Medicine, Yonsei University Wonju College of Medicine, Wonju, Korea
- KMID: 2536970
- DOI: http://doi.org/10.36011/cpp.2020.2.e3
Abstract
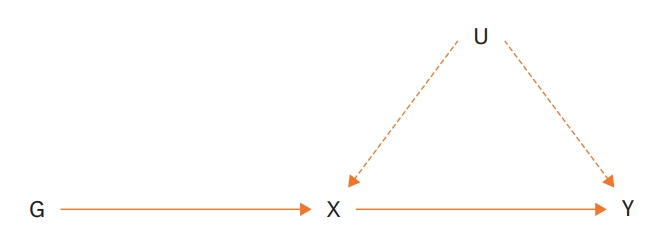
- The Mendelian Randomization (MR) approach is a method that enables causal inference in observational studies. There are 3 assumptions that must be satisfied to obtain suitable results: 1) The genetic variant is strongly associated with the exposure, 2) The genetic variant is independent of the outcome, given the exposure and all confounders (measured and unmeasured) of the exposure-outcome association, 3) The genetic variant is independent of factors (measured and unmeasured) that confound the exposure-outcome relationship. This analysis has been used increasingly since 2011, but many researchers still do not know how to perform MR. Here, we introduce the basic concepts, assumptions, and methods of MR analysis to enable better understanding of this approach.
Keyword
Figure
Reference
-
1. Sekula P, Del Greco M F, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016; 27:3253–65.
Article2. Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, Evans DM, Smith GD. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep. 2017; 4:330–45.
Article3. Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. 1986. Int J Epidemiol. 2004; 33:9.4. Wells D. Mendelian randomisation: a minireview. Winnower. 2015; 2015:3073.
Article5. Burgess S, Thompson S. Mendelian Randomization. New York, NY: Chapman and Hall/CRC;2015.6. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017; 318:1925–6.
Article7. Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003; 32:1–22.
Article8. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008; 27:1133–63.
Article9. Teumer A. Common methods for performing Mendelian randomization. Front Cardiovasc Med. 2018; 5:51.
Article10. Baum C, Schaffer M, Stillman S. Instrumental variables and GMM: estimation and testing. Stata J. 2003; 3:1–31.
Article11. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017; 26:2333–55.
Article12. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011; 40:740–52.
Article13. Angrist JD, Pischke JS. Mostly Harmless Econometrics: An Empiricist's Companion. Princeton, NJ: Princeton University Press;2009.14. Foster EM. Instrumental variables for logistic regression: an illustration. Soc Sci Res. 1997; 26:487–504.
Article15. Angrist JD, Pischke JS. Instrumental variables in action: sometimes you get what you need. In : Angrist JD, Pischke JS, editors. Mostly Harmless Econometrics: An Empiricist's Companion. Princeton, NJ: Princeton University Press;2009. p. 113–220.16. Burgess S, Thompson SG; CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011; 40:755–64.
Article17. Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016; 103:965–78.
Article18. Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007; 16:309–30.
Article19. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018; 362:k601.
Article20. Staiger D, Stock JH. Instrumental variables regression with weak instruments. Econometrica. 1997; 65:557–86.
Article21. Nordestgaard AT, Nordestgaard BG. Coffee intake, cardiovascular disease and all-cause mortality: observational and Mendelian randomization analyses in 95 000-223 000 individuals. Int J Epidemiol. 2016; 45:1938–52.22. Palmer TM, Sterne JA, Harbord RM, Lawlor DA, Sheehan NA, Meng S, Granell R, Smith GD, Didelez V. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am J Epidemiol. 2011; 173:1392–403.
Article23. Wehby GL, Ohsfeldt RL, Murray JC. ‘Mendelian randomization’ equals instrumental variable analysis with genetic instruments. Stat Med. 2008; 27:2745–9.
Article24. Jung KJ, Kim S, Yun M, Jeon C, Jee SH. Review on genetic risk score and cancer prediction models. J Health Info Stat. 2014; 39:1–15.25. Johnson T. Efficient Calculation for Multi-SNP Genetic Risk Scores. American Society of Human Genetics Annual Meeting 2012.26. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014; 23:R89–98.
Article27. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG; EPIC- InterAct Consortium. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015; 30:543–52.
Article28. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018; 7:e34408.
Article29. Timpson NJ, Nordestgaard BG, Harbord RM, Zacho J, Frayling TM, Tybjærg-Hansen A, Smith GD. C-reactive protein levels and body mass index: elucidating direction of causation through reciprocal Mendelian randomization. Int J Obes. 2011; 35:300–8.
Article30. Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986; 1:507–8.
Article31. Wang LN, Zhang Z. Mendelian randomization approach, used for causal inferences. Zhonghua Liu Xing Bing Xue Za Zhi. 2017; 38:547–52.32. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015; 44:512–25.
Article33. Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016; 45:1961–74.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Randomization in clinical studies
- A Review of Mendelian Randomization: Assumptions, Methods, and Application to Obesity-Related Diseases
- Mendelian Randomization Analysis in Observational Epidemiology
- Precision Medicine and Cardiovascular Health: Insights from Mendelian Randomization Analyses
- Exploration of errors in variance caused by using the first-order approximation in Mendelian randomization