J Pathol Transl Med.
2022 Sep;56(5):249-259. 10.4132/jptm.2022.06.11.
Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
- Affiliations
-
- 1Department of Pathology and Translational Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- 3Artificial Intelligence Institute, Seoul National University, Seoul, Korea
- KMID: 2533701
- DOI: http://doi.org/10.4132/jptm.2022.06.11
Abstract
- Background
Activating mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) are predictive biomarkers for response to EGFR–tyrosine kinase inhibitor (TKI) therapy in lung adenocarcinoma (LUAD). Here, we characterized the clinicopathologic features associated with EGFR mutations via peptide nucleic acid clamping-assisted fluorescence melting curve analysis (PANAMutyper) and evaluated the feasibility of targeted deep sequencing for detecting the mutations.
Methods
We examined EGFR mutations in exons 18 through 21 for 2,088 LUADs from July 2017 to April 2020 using PANAMutyper. Of these, we performed targeted deep sequencing in 73 patients and evaluated EGFR-mutation status and TKI clinical response.
Results
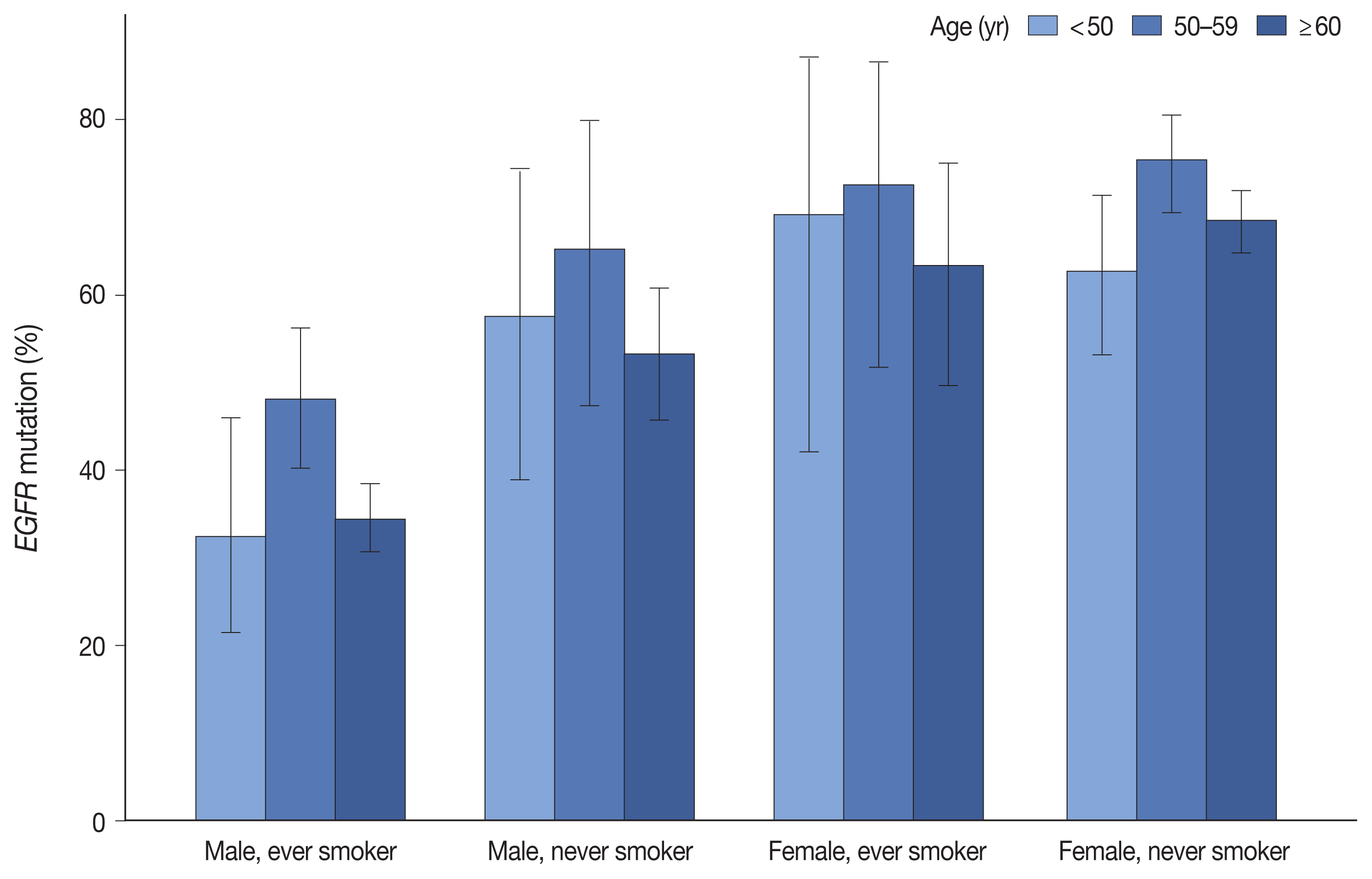
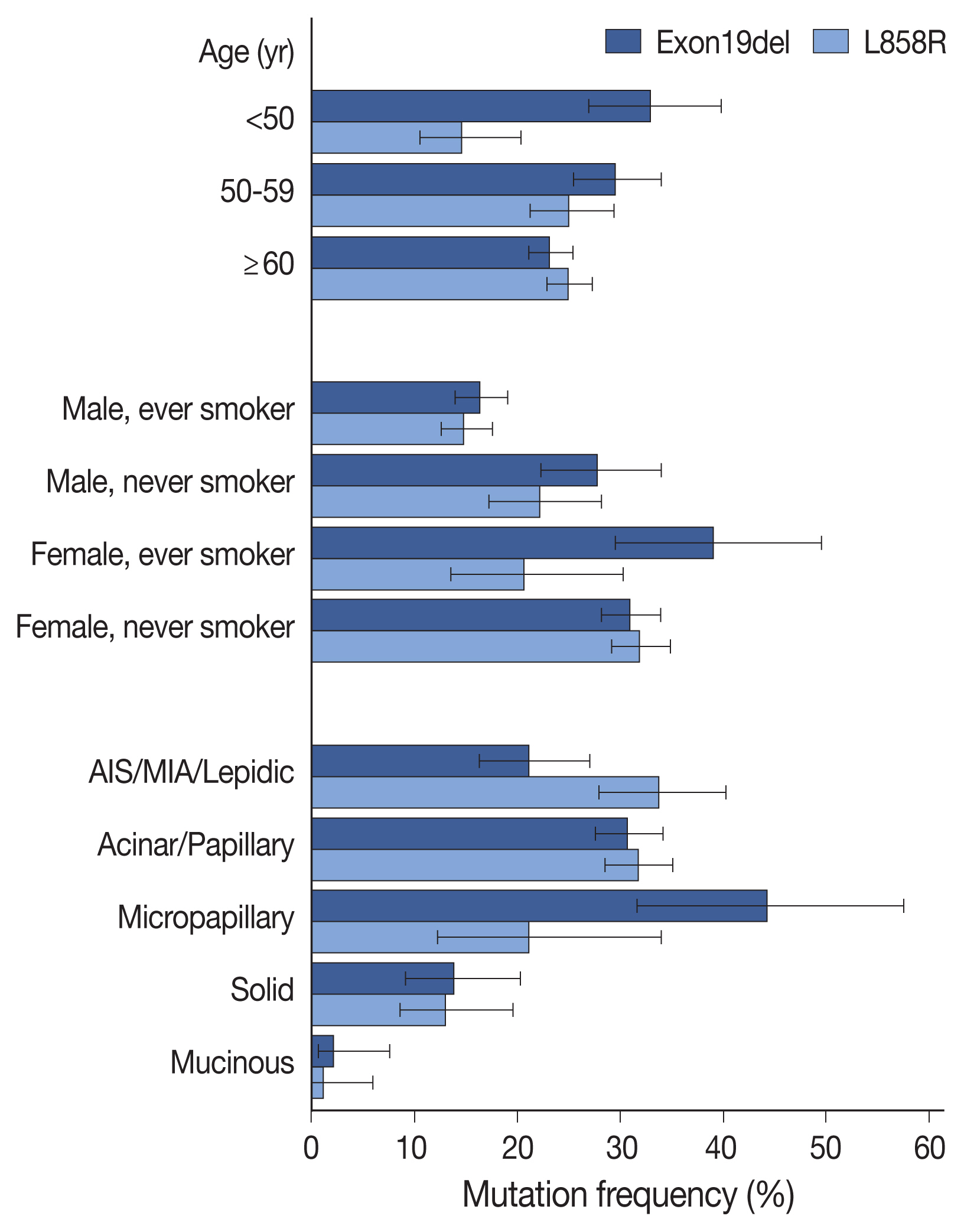
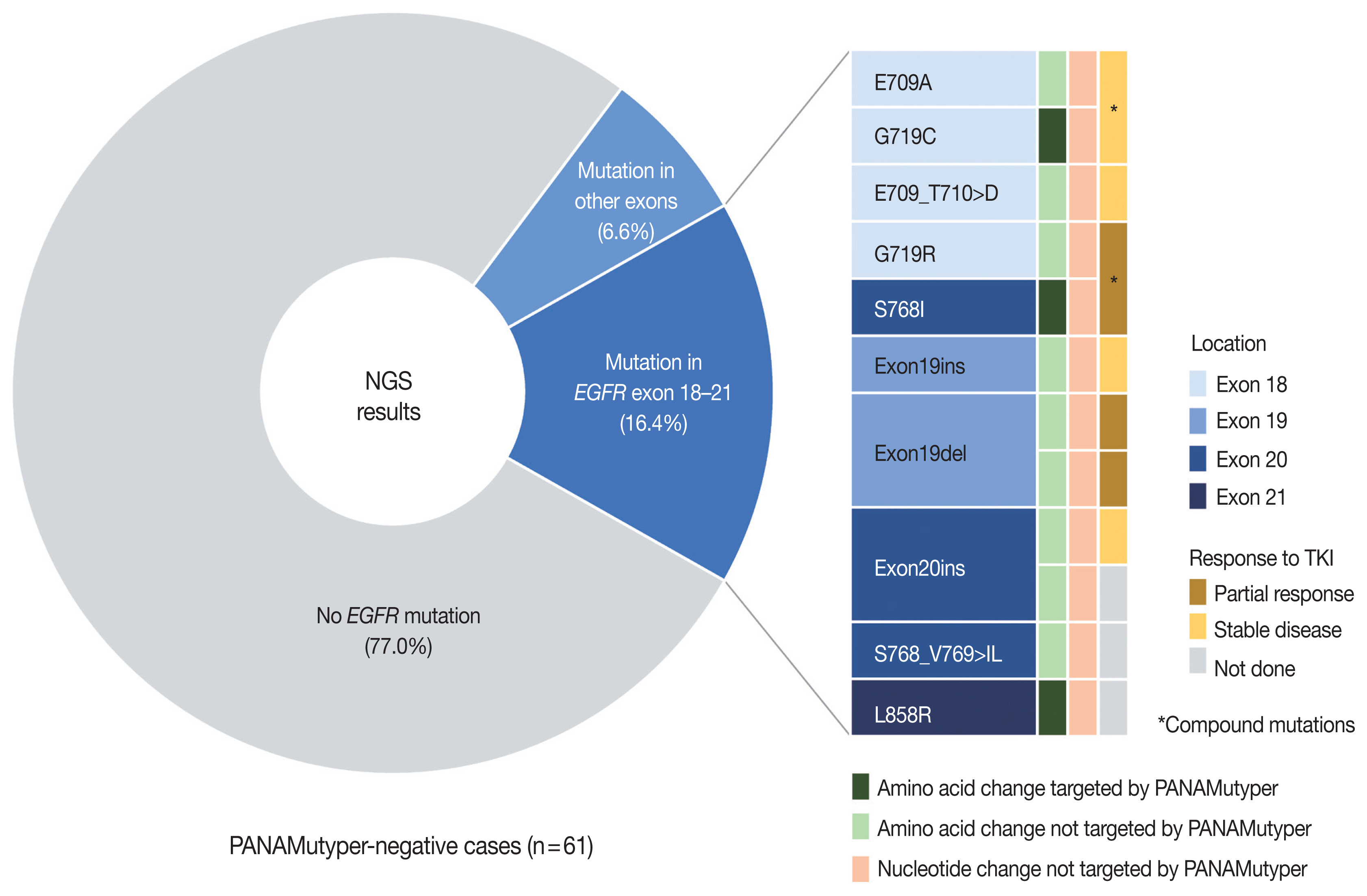
EGFR mutation was identified in 55.7% of LUADs by PANAMutyper, with mutation rates higher in females (69.3%) and never smokers (67.1%) and highest in the age range of 50 to 59 years (64.9%). For the 73 patients evaluated using both methods, next-generation sequencing (NGS) identified EGFR mutation–positive results in 14 of 61 patients (23.0%) who were EGFR-negative according to PANAMutyper testing. Of the 10 patients reportedly harboring a sensitizing mutation according to NGS, seven received TKI treatment, with all showing partial response or stable disease. In the 12 PANAMutyper-positive cases, NGS identified two additional mutations in exon 18, whereas a discordant negative result was observed in two cases.
Conclusions
Although PANAMutyper identified high frequencies of EGFR mutations, targeted deep sequencing revealed additional uncommon EGFR mutations. These findings suggested that appropriate use of NGS may benefit LUAD patients with otherwise negative screening test results.
Figure
Reference
-
References
1. Shim HS, Choi YL, Kim L, et al. Molecular testing of lung cancers. J Pathol Transl Med. 2017; 51:242–54.
Article2. Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015; 10:438–45.3. Lee SH, Kim WS, Choi YD, et al. Analysis of mutations in epidermal growth factor receptor gene in Korean patients with non-small cell lung cancer: summary of a nationwide survey. J Pathol Transl Med. 2015; 49:481–8.
Article4. Kim YT, Seong YW, Jung YJ, et al. The presence of mutations in epidermal growth factor receptor gene is not a prognostic factor for long-term outcome after surgical resection of non-small-cell lung cancer. J Thorac Oncol. 2013; 8:171–8.
Article5. Sun PL, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012; 7:323–30.6. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018; 142:321–46.
Article7. Chang S, Shim HS, Kim TJ, et al. Molecular biomarker testing for non-small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group. J Pathol Transl Med. 2021; 55:181–91.
Article8. Park E, Shim HS. Detection of targetable genetic alterations in Korean lung cancer patients: a comparison study of single-gene assays and targeted next-generation sequencing. Cancer Res Treat. 2020; 52:543–51.
Article9. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018; 13:323–58.10. Han JY, Choi JJ, Kim JY, Han YL, Lee GK. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer. 2016; 16:627.11. Sacher AG, Dahlberg SE, Heng J, Mach S, Janne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2016; 2:313–20.
Article12. Suidan AM, Roisman L, Belilovski Rozenblum A, et al. Lung cancer in young patients: higher rate of driver mutations and brain involvement, but better survival. J Glob Oncol. 2019; 5:1–8.
Article13. Wu SG, Chang YL, Yu CJ, Yang PC, Shih JY. Lung adenocarcinoma patients of young age have lower EGFR mutation rate and poorer efficacy of EGFR tyrosine kinase inhibitors. ERJ Open Res. 2017; 3:00092–2016.14. Evans M, O’Sullivan B, Smith M, et al. Large-scale EGFR mutation testing in clinical practice: analysis of a series of 18,920 non-small cell lung cancer cases. Pathol Oncol Res. 2019; 25:1401–9.15. Tanaka K, Hida T, Oya Y, et al. Unique prevalence of oncogenic genetic alterations in young patients with lung adenocarcinoma. Cancer. 2017; 123:1731–40.
Article16. Zhang Y, He D, Fang W, et al. The difference of clinical characteristics between patients with exon 19 deletion and those with L858R mutation in nonsmall cell lung cancer. Medicine (Baltimore). 2015; 94:e1949.
Article17. Chen Z, Zhang J, Huang K, Shen Q, Teng X. Comparison of clinicopathologic characteristics between patients with EGFR exon 19 deletion and EGFR L858R mutation in lung cancer. Int J Clin Exp Pathol. 2018; 11:4644–9.18. Lee B, Lee T, Lee SH, Choi YL, Han J. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget. 2016; 7:23874–84.
Article19. Kadota K, Yeh YC, D’Angelo SP, et al. Associations between mutations and histologic patterns of mucin in lung adenocarcinoma: invasive mucinous pattern and extracellular mucin are associated with KRAS mutation. Am J Surg Pathol. 2014; 38:1118–27.20. Shim HS, Kenudson M, Zheng Z, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J Thorac Oncol. 2015; 10:1156–62.
Article21. Shim HS, Lee DH, Park EJ, Kim SH. Histopathologic characteristics of lung adenocarcinomas with epidermal growth factor receptor mutations in the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification. Arch Pathol Lab Med. 2011; 135:1329–34.
Article22. Lu F, Li S, Dong B, Zhang S, Lv C, Yang Y. Identification of lung adenocarcinoma mutation status based on histologic subtype: retrospective analysis of 269 patients. Thorac Cancer. 2016; 7:17–23.
Article23. Li Y, Tan Y, Hu S, et al. Targeted sequencing analysis of predominant histological subtypes in resected stage I invasive lung adenocarcinoma. J Cancer. 2021; 12:3222–9.
Article24. Yoo SB, Chung JH, Lee HJ, Lee CT, Jheon S, Sung SW. Epidermal growth factor receptor mutation and p53 overexpression during the multistage progression of small adenocarcinoma of the lung. J Thorac Oncol. 2010; 5:964–9.
Article25. Jia M, Yu S, Cao L, Sun PL, Gao H. Clinicopathologic features and genetic alterations in adenocarcinoma in situ and minimally invasive adenocarcinoma of the lung: long-term follow-up study of 121 Asian patients. Ann Surg Oncol. 2020; 27:3052–63.
Article26. Ishida H, Shimizu Y, Sakaguchi H, et al. Distinctive clinicopathological features of adenocarcinoma in situ and minimally invasive adenocarcinoma of the lung: a retrospective study. Lung Cancer. 2019; 129:16–21.
Article27. Caso R, Sanchez-Vega F, Tan KS, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. 2020; 15:1844–56.
Article28. Tavernari D, Battistello E, Dheilly E, et al. Nongenetic evolution drives lung adenocarcinoma spatial heterogeneity and progression. Cancer Discov. 2021; 11:1490–507.
Article29. Sun PL, Jin Y, Kim H, Lee CT, Jheon S, Chung JH. High concordance of EGFR mutation status between histologic and corresponding cytologic specimens of lung adenocarcinomas. Cancer Cytopathol. 2013; 121:311–9.30. Ku BM, Heo MH, Kim JH, et al. Molecular screening of small biopsy samples using next-generation sequencing in Korean patients with advanced non-small cell lung cancer: Korean Lung Cancer Consortium (KLCC-13-01). J Pathol Transl Med. 2018; 52:148–56.
Article31. Oztan A, Fischer S, Schrock AB, et al. Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer. 2017; 111:84–7.
Article32. Brown BP, Zhang YK, Westover D, et al. On-target resistance to the mutant-selective EGFR inhibitor osimertinib can develop in an allele-specific manner dependent on the original EGFR-activating mutation. Clin Cancer Res. 2019; 25:3341–51.33. Fassunke J, Muller F, Keul M, et al. Overcoming EGFR(G724S)-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun. 2018; 9:4655.34. Nukaga S, Yasuda H, Tsuchihara K, et al. Amplification of EGFR wild-type alleles in non-small cell lung cancer cells confers acquired resistance to mutation-selective EGFR tyrosine kinase inhibitors. Cancer Res. 2017; 77:2078–89.35. Knebel FH, Bettoni F, Shimada AK, et al. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer. 2017; 108:238–41.
Article36. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019; 121:725–37.
Article37. Canale M, Petracci E, Delmonte A, et al. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res. 2017; 23:2195–202.38. Roeper J, Falk M, Chalaris-Rissmann A, et al. TP53 co-mutations in EGFR mutated patients in NSCLC stage IV: a strong predictive factor of ORR, PFS and OS in EGFR mt+ NSCLC. Oncotarget. 2020; 11:250–64.
Article39. Blakely CM, Watkins TB, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017; 49:1693–704.
Article40. Liu Q, Yu S, Zhao W, Qin S, Chu Q, Wu K. EGFR-TKIs resistance via EGFR-independent signaling pathways. Mol Cancer. 2018; 17:53.
Article41. Kohsaka S, Petronczki M, Solca F, Maemondo M. Tumor clonality and resistance mechanisms in EGFR mutation-positive non-small-cell lung cancer: implications for therapeutic sequencing. Future Oncol. 2019; 15:637–52.42. Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019; 19:495–509.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- New Lung Cancer Panel for High-Throughput Targeted Resequencing
- Morule-like features in pulmonary adenocarcinoma associated with epidermal growth factor receptor mutations: two case reports with targeted next-generation sequencing analysis
- Detection of Targetable Genetic Alterations in Korean Lung CancerPatients: A Comparison Study of Single-Gene Assays andTargeted Next-Generation Sequencing
- A case of concomitant EGFR/ALK alteration against a mutated EGFR background in early-stage lung adenocarcinoma
- Comparison of Clinicopathogenomic Features and Treatment Outcomes of EGFR and HER2 Exon 20 Insertion Mutations in Non–Small Cell Lung Cancer: Single-Institution Experience