Detection of Targetable Genetic Alterations in Korean Lung CancerPatients: A Comparison Study of Single-Gene Assays andTargeted Next-Generation Sequencing
- Affiliations
-
- 1Department of Pathology, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2500339
- DOI: http://doi.org/10.4143/crt.2019.305
Abstract
- Purpose
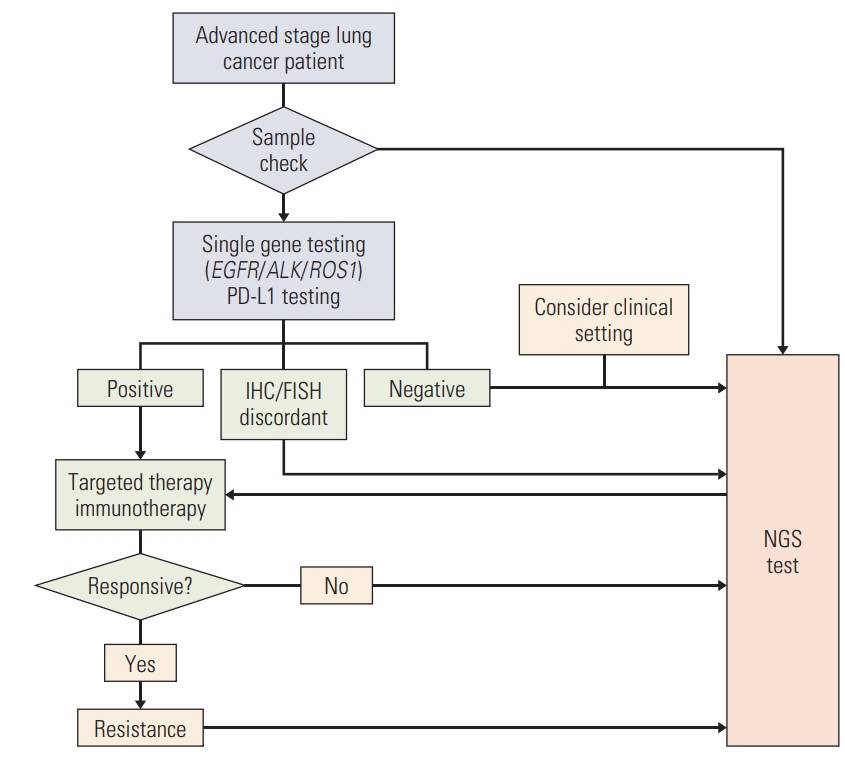
Epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), and ROS proto-oncogene 1 (ROS1) are ‘must-test’ biomarkers in the molecular diagnostics of advanced- stage lung cancer patients. Although single-gene assays are currently considered the gold standard for these genes, next-generation sequencing (NGS) tests are being introduced to clinical practices. We compared the results of current diagnostics and aimed to suggest timely effective guidance for their clinical use.
Materials and Methods
Patients with lung cancer who received both conventional single-gene assays and subsequent targeted NGS testing were enrolled, and the results of their tests were compared.
Results
A total of 241 patients were enrolled, and the EGFR real-time polymerase chain reaction, ALK fluorescence in situ hybridization (FISH), and ROS1 FISH assays exhibited 92.9%, 99.6%, and 99.5% concordance with the NGS tests, respectively. The discordant cases were mostly false-negatives of the single-gene assays, probably due to technical limitation. Of 158 cases previously designated as wild-type, EGFR, ALK, and ROS1 alterations were identified in 10.1%, 1.9%, and 1.3%, respectively, and other targetable alterations were identified in 36.1% of the cases. Of patients with additionally identified actionable alterations, 32.6% (31/95) received matched therapy with a clinical benefit of 48.4% (15/31).
Conclusion
Even though the conventional and NGS methods were concordant in the majority of cases, NGS testing still revealed a considerable number of additional EGFR, ALK, and ROS1 alterations, as well as other targetable alterations, in Korean advanced-stage lung cancer patients. Given the high frequency of EGFR and other targetable mutations identified in the present study, NGS testing is highly recommended in the diagnosis of Korean lung cancer patients.
Keyword
Figure
Cited by 3 articles
-
Increased Radiosensitivity of Solid Tumors Harboring
ATM andBRCA1/2 Mutations
Kyung Hwan Kim, Han Sang Kim, Seung-seob Kim, Hyo Sup Shim, Andrew Jihoon Yang, Jason Joon Bock Lee, Hong In Yoon, Joong Bae Ahn, Jee Suk Chang
Cancer Res Treat. 2022;54(1):54-64. doi: 10.4143/crt.2020.1247.Comparison of the Data of a Next-Generation Sequencing Panel from K-MASTER Project with That of Orthogonal Methods for Detecting Targetable Genetic Alterations
Yoon Ji Choi, Jung Yoon Choi, Ju Won Kim, Ah Reum Lim, Youngwoo Lee, Won Jin Chang, Soohyeon Lee, Jae Sook Sung, Hee-Joon Chung, Jong Won Lee, Eun Joo Kang, Jung Sun Kim, Taekyu Lim, Hye Sook Kim, Yu Jung Kim, Mi Sun Ahn, Young Saing Kim, Ji Hyun Park, Seungtaek Lim, Sung Shim Cho, Jang Ho Cho, Sang Won Shin, Kyong Hwa Park, Yeul Hong Kim
Cancer Res Treat. 2022;54(1):30-39. doi: 10.4143/crt.2021.218.Landscape of
EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
J Pathol Transl Med. 2022;56(5):249-259. doi: 10.4132/jptm.2022.06.11.
Reference
-
References
1. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018; 13:323–58.
Article2. Shim HS, Choi YL, Kim L, Chang S, Kim WS, Roh MS, et al. Molecular testing of lung cancers. J Pathol Transl Med. 2017; 51:242–54.
Article3. Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017; 7:596–609.
Article4. Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015; 121:631–9.
Article5. Pennell NA, Mutebi A, Zhou ZY, Ricculli ML, Tang W, Wang H, et al. Economic impact of next generation sequencing vs sequential single-gene testing modalities to detect genomic alterations in metastatic non-small cell lung cancer using a decision analytic model. J Clin Oncol. 2018; 36(15 Suppl):9031.
Article6. Shao D, Lin Y, Liu J, Wan L, Liu Z, Cheng S, et al. A targeted next-generation sequencing method for identifying clinically relevant mutation profiles in lung adenocarcinoma. Sci Rep. 2016; 6:22338.
Article7. Cha YJ, Lee JS, Kim HR, Lim SM, Cho BC, Lee CY, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One. 2014; 9:e103333.8. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23.9. Legras A, Barritault M, Tallet A, Fabre E, Guyard A, Rance B, et al. Validity of targeted next-generation sequencing in routine care for identifying clinically relevant molecular profiles in non-small-cell lung cancer: results of a 2-year experience on 1343 samples. J Mol Diagn. 2018; 20:550–64.10. Yu G, Xie X, Sun D, Geng J, Fu F, Zhang L, et al. EGFR mutation L747P led to gefitinib resistance and accelerated liver metastases in a Chinese patient with lung adenocarcinoma. Int J Clin Exp Pathol. 2015; 8:8603–6.11. Wu SG, Gow CH, Yu CJ, Chang YL, Yang CH, Hsu YC, et al. Frequent epidermal growth factor receptor gene mutations in malignant pleural effusion of lung adenocarcinoma. Eur Respir J. 2008; 32:924–30.
Article12. Tuononen K, Maki-Nevala S, Sarhadi VK, Wirtanen A, Ronty M, Salmenkivi K, et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer. 2013; 52:503–11.13. Kauffmann-Guerrero D, Huber RM, Reu S, Tufman A, Mertsch P, Syunyaeva Z, et al. NSCLC patients harbouring rare or complex EGFR mutations are more often smokers and might not benefit from first-line tyrosine kinase inhibitor therapy. Respiration. 2018; 95:169–76.14. Tu HY, Ke EE, Yang JJ, Sun YL, Yan HH, Zheng MY, et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer. 2017; 114:96–102.15. Kobayashi Y, Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 2016; 107:1179–86.
Article16. Vyse S, Huang PH. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct Target Ther. 2019; 4:5.
Article17. Voon PJ, Tsui DW, Rosenfeld N, Chin TM. EGFR exon 20 insertion A763-Y764insFQEA and response to erlotinib: letter. Mol Cancer Ther. 2013; 12:2614–5.18. Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med. 2018; 24:638–46.19. Rosenbaum JN, Bloom R, Forys JT, Hiken J, Armstrong JR, Branson J, et al. Genomic heterogeneity of ALK fusion breakpoints in non-small-cell lung cancer. Mod Pathol. 2018; 31:791–808.20. Ali SM, Hensing T, Schrock AB, Allen J, Sanford E, Gowen K, et al. Comprehensive genomic profiling identifies a subset of crizotinib-responsive ALK-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist. 2016; 21:762–70.21. Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L, Ilouze M, Dvir A, Boyle T, et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist. 2015; 20:316–22.22. van der Wekken AJ, Pelgrim R, 't Hart N, Werner N, Mastik MF, Hendriks L, et al. Dichotomous ALK-IHC is a better predictor for ALK inhibition outcome than traditional ALK-FISH in advanced non-small cell lung cancer. Clin Cancer Res. 2017; 23:4251–8.23. Cabillic F, Hofman P, Ilie M, Peled N, Hochmair M, Dietel M, et al. ALK IHC and FISH discordant results in patients with NSCLC and treatment response: for discussion of the question-to treat or not to treat? ESMO Open. 2018; 3:e000419.24. Lin C, Shi X, Yang S, Zhao J, He Q, Jin Y, et al. Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non-small-cell lung cancer. Lung Cancer. 2019; 131:62–8.25. Choi CM, Kim HC, Jung CY, Cho DG, Jeon JH, Lee JE, et al. Report of the Korean Association of Lung Cancer Registry (KALC-R), 2014. Cancer Res Treat. 2019; 51:1400–10.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Validation and Clinical Application of ONCOaccuPanel for Targeted Next-Generation Sequencing of Solid Tumors
- Integrating a Next Generation Sequencing Panel into Clinical Practice in Ovarian Cancer
- Recent Advances in the Clinical Application of Next-Generation Sequencing
- Principles of Genetic Counseling in the Era of Next-Generation Sequencing
- Detection of Mosaic Sequence Variants Associated with Human Genetic Diseases