Yonsei Med J.
2019 Oct;60(10):914-923. 10.3349/ymj.2019.60.10.914.
Integrating a Next Generation Sequencing Panel into Clinical Practice in Ovarian Cancer
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Institute of Women's Life Medical Science, Yonsei University College of Medicine, Seoul, Korea. jungyunlee@yuhs.ac
- 2Department of Pathology, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2459144
- DOI: http://doi.org/10.3349/ymj.2019.60.10.914
Abstract
- PURPOSE
Few efforts have been made to integrate a next generation sequencing (NGS) panel into standard clinical treatment of ovarian cancer. The aim of this study was to investigate the clinical utility of NGS and to identify clinically impactful information beyond targetable alterations.
MATERIALS AND METHODS
We conducted a retrospective review of 84 patients with ovarian cancer who underwent NGS between March 1, 2017, and July 31, 2018, at the Yonsei Cancer Hospital. We extracted DNA from formalin-fixed, paraffin-embedded tissue samples of ovarian cancer. The TruSight Tumor 170 gene panel was used to prepare libraries, and the MiSeq instrument was used for NGS.
RESULTS
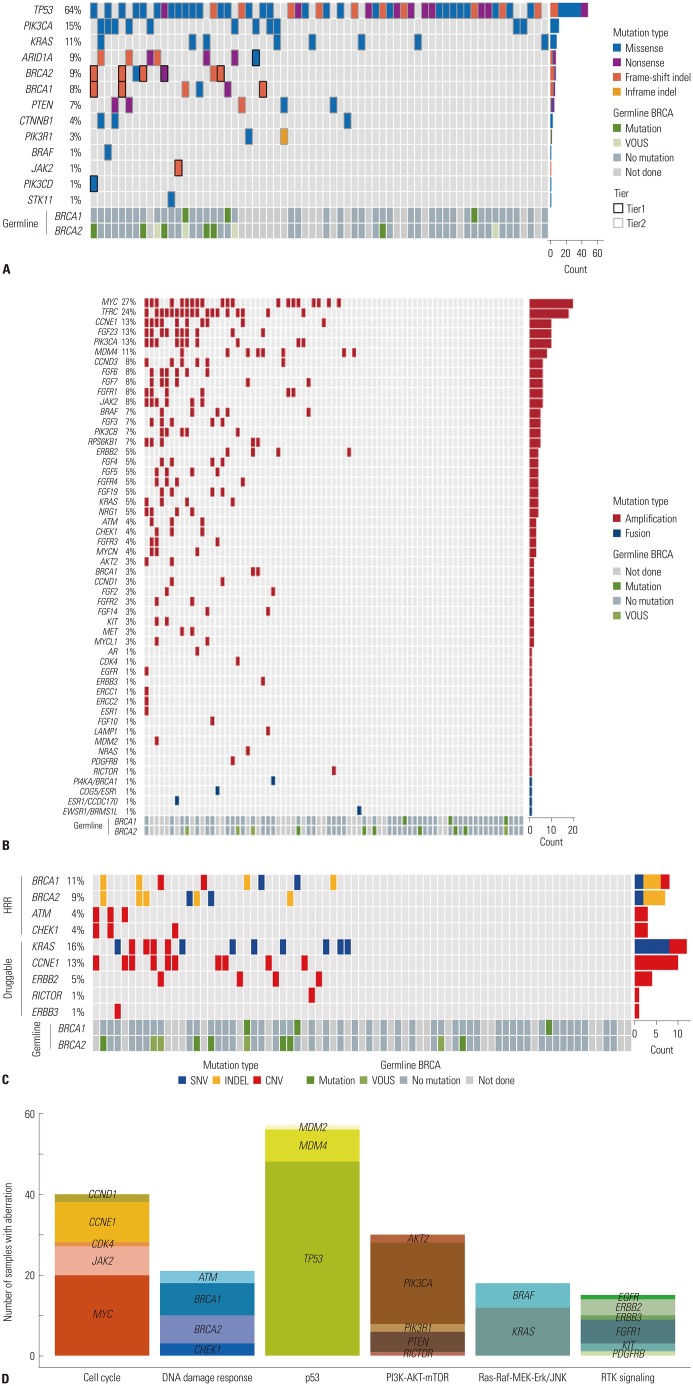
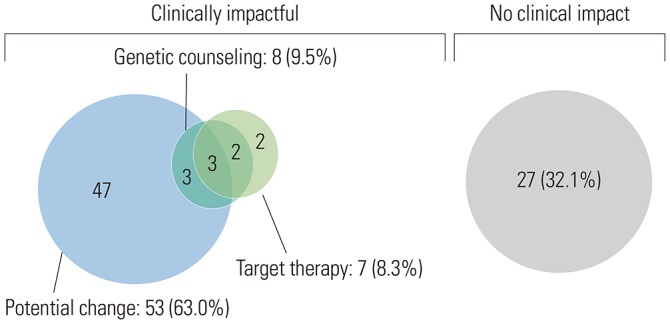
Of the 84 patients, 55 (65.1%) had high-grade serous carcinomas. Seventy-three (86.7%) patients underwent NGS at the time of diagnosis, and 11 (13.3%) underwent NGS upon relapse. The most common genetic alterations were in TP53 (64%), PIK3CA (15%), and BRCA1/2 (13%), arising as single nucleotide variants and indels. MYC amplification (27%) was the most common copy number variation and fusion. Fifty-seven (67.9%) patients had more than one actionable alteration other than TP53. Seven (8.3%) cases received matched-target therapy based on the following sequencing results: BRCA1 or 2 mutation, poly ADP ribose polymerase inhibitor (n=5); PIK3CA mutation, AKT inhibitor (n=1); and MLH1 mutation, PD-1 inhibitor (n=1). Fifty-three (63.0%) patients had a possibility of treatment change, and 8 (9.5%) patients received genetic counseling.
CONCLUSION
Implementation of NGS may help in identifying patients who might benefit from targeted treatment therapies and genetic counseling.
MeSH Terms
Figure
Reference
-
1. Lee JY, Kim EY, Jung KW, Shin A, Chan KK, Aoki D, et al. Trends in gynecologic cancer mortality in East Asian regions. J Gynecol Oncol. 2014; 25:174–182. PMID: 25045429.
Article2. Lim MC, Moon EK, Shin A, Jung KW, Won YJ, Seo SS, et al. Incidence of cervical, endometrial, and ovarian cancer in Korea, 1999-2010. J Gynecol Oncol. 2013; 24:298–302. PMID: 24167664.
Article3. Winter WE 3rd, Maxwell GL, Tian C, Carlson JW, Ozols RF, Rose PG, et al. Prognostic factors for stage III epithelial ovarian cancer: a Gynecologic Oncology Group Study. J Clin Oncol. 2007; 25:3621–3627. PMID: 17704411.
Article4. Groisberg R, Hong DS, Roszik J, Janku F, Tsimberidou AM, Javle M, et al. Clinical next-generation sequencing for precision oncology in rare cancers. Mol Cancer Ther. 2018; 17:1595–1601. PMID: 29654067.
Article5. Carr TH, McEwen R, Dougherty B, Johnson JH, Dry JR, Lai Z, et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer. 2016; 16:319–329. PMID: 27112209.
Article6. Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y. Integrated genomic characterization of endometrial carcinoma. Nature. 2013; 497:67–73. PMID: 23636398.
Article7. Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013; 45:1113–1120. PMID: 24071849.
Article8. Fisher KE, Zhang L, Wang J, Smith GH, Newman S, Schneider TM, et al. Clinical validation and implementation of a targeted next-generation sequencing assay to detect somatic variants in non-small cell lung, melanoma, and gastrointestinal malignancies. J Mol Diagn. 2016; 18:299–315. PMID: 26801070.
Article9. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23. PMID: 27993330.10. Joseph L, Cankovic M, Caughron S, Chandra P, Emmadi R, Hagenkord J, et al. The spectrum of clinical utilities in molecular pathology testing procedures for inherited conditions and cancer: a report of the Association for Molecular Pathology. J Mol Diagn. 2016; 18:605–619. PMID: 27542512.11. Oberg JA, Glade Bender JL, Sulis ML, Pendrick D, Sireci AN, Hsiao SJ, et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med. 2016; 8:133. PMID: 28007021.
Article12. Heong V, Syn NL, Lee XW, Sapari NS, Koh XQ, Adam Isa ZF, et al. Value of a molecular screening program to support clinical trial enrollment in Asian cancer patients: the Integrated Molecular Analysis of Cancer (IMAC) Study. Int J Cancer. 2018; 142:1890–1900. PMID: 28994108.
Article13. Le Tourneau C, Delord JP, Gonçalves A, Gavoille C, Dubot C, Isambert N, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015; 16:1324–1334. PMID: 26342236.
Article14. Loeb KR, Loeb LA. Significance of multiple mutations in cancer. Carcinogenesis. 2000; 21:379–385. PMID: 10688858.
Article15. Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011; 29:3008–3015. PMID: 21709188.
Article16. Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015; 521:489–494. PMID: 26017449.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Validity of Next-Generation Sequencing Multi-Gene Panel Testing for Detecting Pathogenic Variants in Patients With Hereditary Breast-Ovarian Cancer Syndrome
- Utility of Next-Generation Sequencing Panel Including Hereditary Breast and Ovarian Cancer-Related Genes for Pathogenic Variant Detection
- Genetic tests by next-generation sequencing in children with developmental delay and/or intellectual disability
- Development of an RNA sequencing panel to detect gene fusions in thyroid cancer
- Role and clinical application of next-generation sequencing (NGS) for ovarian cancer