Blood Res.
2021 Dec;56(4):252-258. 10.5045/br.2021.2021016.
Mutation analysis and characterisation of F9 gene in haemophilia- B population of India
- Affiliations
-
- 1Human Genetics Laboratory, Department of Anatomy, Shri B.M Patil Medical College, Hospital and Research Centre, BLDE (Deemed to be University), Vijayapura, India
- 2Division of Human Genetics (Central Research Lab), Bagalkot, India
- 3Department of Anatomy, S. Nijaliangappa Medical College, HSK Hospital and Research Center, Bagalkot, India
- 4Karnataka Institute for DNA Research (KIDNAR), Dharwad, Karnataka, India
- 5Department of Pathology, J. J. M. Medical College, Davangere, Karnataka, India
- 6Laboratory of Vascular Physiology and Medicine, Department of Physiology, Shri B. M. Patil Medical College, Hospital and Research Centre, BLDE (Deemed to be University), Vijayapura, India
- KMID: 2524077
- DOI: http://doi.org/10.5045/br.2021.2021016
Abstract
- Background
Hemophilia B (HB) is an X-linked bleeding disorder resulting from coagulation factor IX defects. Over 3,000 pathogenic, HB-associated mutations in the F9 gene have been identified. We aimed to investigate the role of F9 variants in 150 HB patients using sequencing technology.
Methods
F9 gene sequences were amplified from peripheral blood-derived DNA and sequenced on an Applied Biosystems (ABI) 3500 Sanger sequencing platform. Functional and structural predictions of mutant FIX were analyzed.
Results
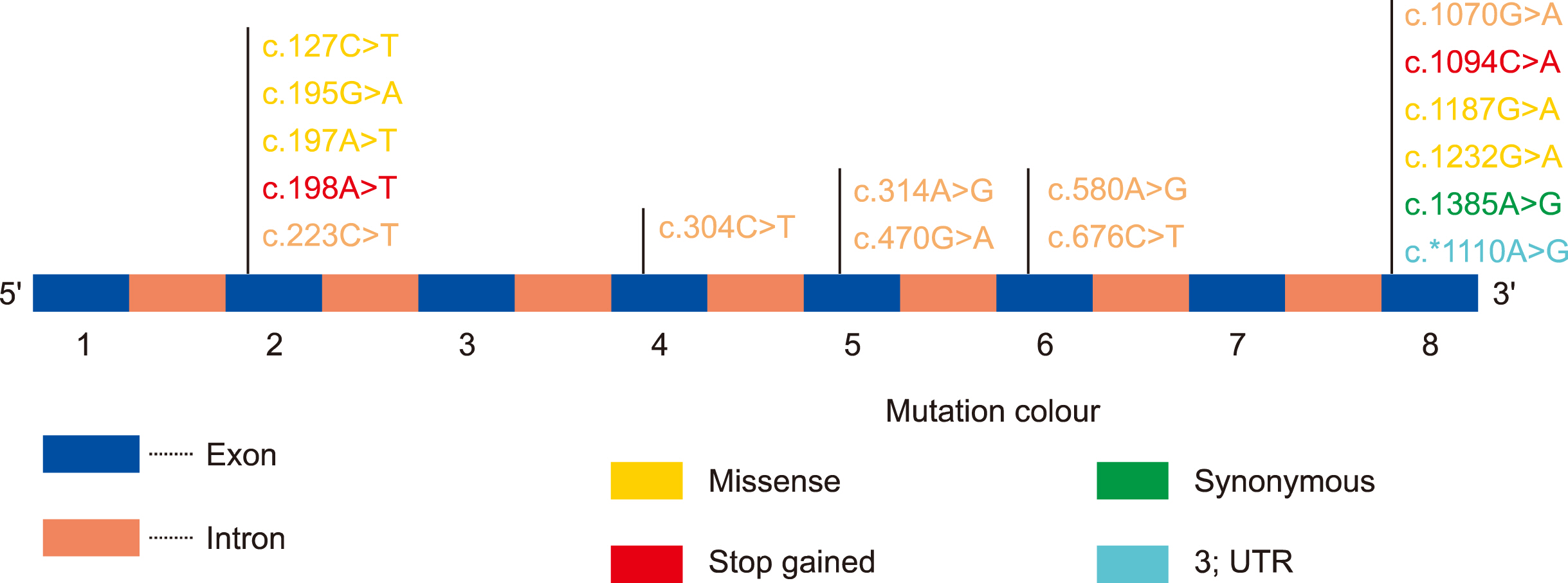
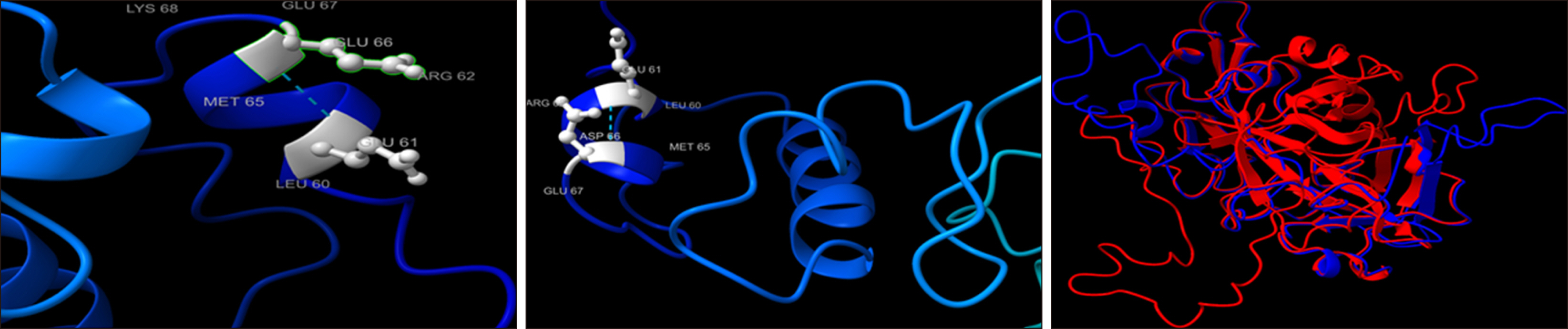
Among 150 HB patients, 102 (68%), 30 (20%), and 18 (12%) suffered from severe, moderate, and mild HB, respectively. Genetic analysis identified 16 mutations, including 3 novel mutations. Nine mutations (7 missense and 2 stop-gain) were found to be pathogenic. Only 3 mutations (c.127C>T, c.470G>A, and c.1070G>A) were associated with different severities. While 2 mutations were associated with mild HB cases (c.304C>T and c.580A>G), 2 (c.195G>A and c.1385A>G) and 3 mutations (c.223C>T, c.1187G>A, and c.1232G>A) resulted in moderate and severe disease, respectively. Additionally, 1 mutation each was associated with mild-moderate (c.*1110A>G) and mild-severe HB disease (c.197A>T), 4 mutations were associated with moderate-severe HB cases (c.314A>G, c.198A>T, c.676C>T, and c.1094C>A). FIX concentrations were lower in the mutated group (5.5±2.5% vs. 8.0±2.5%). Novel p.E66D and p.S365 mutations were predicted to be pathogenic based on changes in FIX structure and function.
Conclusion
Novel single nucleotide polymorphisms (SNPs) largely contributed to the pathogenesis of HB. Our study strongly suggests that population-based genetic screening will be particularly helpful to identify risk prediction and carrier detection tools for Indian HB patients.
Keyword
Figure
Reference
-
1. Yu T, Dai J, Liu H, et al. 2012; Spectrum of F9 mutations in Chinese haemophilia B patients: identification of 20 novel mutations. Pathology. 44:342–7. DOI: 10.1097/PAT.0b013e328353443d. PMID: 22544209.
Article2. Rosendaal FR, Smit C, Briët E. 1991; Hemophilia treatment in historical perspective: a review of medical and social developments. Ann Hematol. 62:5–15. DOI: 10.1007/BF01714977. PMID: 1903310.
Article3. Mannucci PM. 2003; Treatment of hemophilia: recombinant factors only? No. J Thromb Haemost. 1:216–7. DOI: 10.1046/j.1538-7836.2003.00046.x. PMID: 12871490.
Article4. Goodeve AC. 2015; Hemophilia B: molecular pathogenesis and mutation analysis. J Thromb Haemost. 13:1184–95. DOI: 10.1111/jth.12958. PMID: 25851415. PMCID: PMC4496316.5. Parrado Jara YA, Yunis Hazbun LK, Linares A, Yunis Londoño JJ. 2020; Molecular characterization of hemophilia B patients in Colombia. Mol Genet Genomic Med. 8:e1210. DOI: 10.1002/mgg3.1210. PMID: 32155688. PMCID: PMC7216803.
Article6. Yuen LK, Zakaria Z, Yusoff YM, Esa E, Afandi FM, Karim FDA. 2017; A novel missense mutation of F9 gene in hemophilia B Patients. J Blood Disord Transfus. 8:383.7. Bhattacharya DK. 2006; Haemophilia in the Indian Scenario. Int J Hum Genet. 6:33–9. DOI: 10.1080/09723757.2006.11885944.
Article8. Kumar S, Sinha S, Bharti A, Meena LP, Gupta V, Shukla J. 2019; A study to determine the prevalence, clinical profile and incidence of formation of inhibitors in patients of hemophilia in North Eastern part of India. J Family Med Prim Care. 8:2463–7. DOI: 10.4103/jfmpc.jfmpc_316_19. PMID: 31463277. PMCID: PMC6691464.
Article9. Yi S, Yang Q, Zuo Y, et al. 2020; A novel missense mutation in F9 gene causes hemophilia B in a family with clinical variability. Blood Coagul Fibrinolysis. 31:121–6. DOI: 10.1097/MBC.0000000000000884. PMID: 31904612.
Article10. Geddes VA, MacGillivray RT. 1987; The molecular genetics of hemophilia B. Transfus Med Rev. 1:161–70. DOI: 10.1016/S0887-7963(87)70018-2. PMID: 2980275.
Article11. Li T, Miller CH, Payne AB, Craig Hooper W. 2013; The CDC hemophilia B mutation project mutation list: a new online resource. Mol Genet Genomic Med. 1:238–45. DOI: 10.1002/mgg3.30. PMID: 24498619. PMCID: PMC3865591.12. Simhadri VL, Hamasaki-Katagiri N, Lin BC, et al. 2017; Single synonymous mutation in factor IX alters protein properties and underlies haemophilia B. J Med Genet. 54:338–45. DOI: 10.1136/jmedgenet-2016-104072. PMID: 28007939. PMCID: PMC6192418.
Article13. Zahari M, Sulaiman SA, Othman Z, Ayob Y, Karim FA, Jamal R. 2018; Mutational profiles of F8 and F9 in a cohort of haemophilia A and haemophilia B patients in the multi-ethnic Malaysian population. Mediterr J Hematol Infect Dis. 10:e2018056. DOI: 10.4084/mjhid.2018.056. PMID: 30210749. PMCID: PMC6131101.
Article14. Huang L, Li L, Lin S, et al. 2020; Molecular analysis of 76 Chinese hemophilia B pedigrees and the identification of 10 novel mutations. Mol Genet Genomic Med. 8:e1482. DOI: 10.1002/mgg3.1482. PMID: 32875744. PMCID: PMC7667291.
Article15. Abla Z, Mouloud Y, Hejer EM, et al. 2018; Mutations causing hemophilia B in Algeria: identification of two novel mutations of the factor 9 gene. Biodiversitas. 19:52–8. DOI: 10.13057/biodiv/d190109.
Article16. Radic CP, Rossetti LC, Abelleyro MM, et al. 2013; Assessment of the F9 genotype-specific FIX inhibitor risks and characterisation of 10 novel severe F9 defects in the first molecular series of Argentinian patients with haemophilia B. Thromb Haemost. 109:24–33. DOI: 10.1160/TH12-05-0302. PMID: 23093250. PMCID: PMC4220540.
Article17. Kwon MJ, Yoo KY, Kim HJ, Kim SH. 2008; Identification of mutations in the F9 gene including exon deletion by multiplex ligation-dependent probe amplification in 33 unrelated Korean patients with haemophilia B. Haemophilia. 14:1069–75. DOI: 10.1111/j.1365-2516.2008.01796.x. PMID: 18624698.18. Saini S, Hamasaki-Katagiri N, Pandey GS, et al. 2015; Genetic determinants of immunogenicity to factor IX during the treatment of haemophilia B. Haemophilia. 21:210–8. DOI: 10.1111/hae.12553. PMID: 25470321.19. Koeberl DD, Bottema CD, Sarkar G, Ketterling RP, Chen SH, Sommer SS. 1990; Recurrent nonsense mutations at arginine residues cause severe hemophilia B in unrelated hemophiliacs. Hum Genet. 84:387–90. DOI: 10.1007/BF00195805. PMID: 1969838.
Article20. Suehiro K, Kawabata S, Miyata T, et al. 1989; Blood clotting factor IX BM Nagoya. Substitution of arginine 180 by tryptophan and its activation by alpha-chymotrypsin and rat mast cell chymase. J Biol Chem. 264:21257–65. DOI: 10.1016/S0021-9258(19)30074-2. PMID: 2592373.21. Bertina RM, van der Linden IK, Mannucci PM, et al. 1990; Mutations in hemophilia Bm occur at the Arg180-Val activation site or in the catalytic domain of factor IX. J Biol Chem. 265:10876–83. DOI: 10.1016/S0021-9258(19)38528-X. PMID: 2162822.
Article22. Taylor SA, Deugau KV, Lillicrap DP. 1991; Somatic mosaicism and female-to-female transmission in a kindred with hemophilia B (factor IX deficiency). Proc Natl Acad Sci U S A. 88:39–42. DOI: 10.1073/pnas.88.1.39. PMID: 1986380. PMCID: PMC50743.
Article23. Li Y, Bezemer ID, Rowland CM, et al. 2009; Genetic variants associated with deep vein thrombosis: the F11 locus. J Thromb Haemost. 7:1802–8. DOI: 10.1111/j.1538-7836.2009.03544.x. PMID: 19583818.24. Graham JB, Lubahn DB, Lord ST, et al. 1988; The Malmö polymorphism of coagulation factor IX, an immunologic polymorphism due to dimorphism of residue 148 that is in linkage disequilibrium with two other F.IX polymorphisms. Am J Hum Genet. 42:573–80. PMID: 2450455. PMCID: PMC1715231.25. McGraw RA, Davis LM, Noyes CM, et al. 1985; Evidence for a prevalent dimorphism in the activation peptide of human coagulation factor IX. Proc Natl Acad Sci U S A. 82:2847–51. DOI: 10.1073/pnas.82.9.2847. PMID: 3857619. PMCID: PMC397663.
Article26. Miyata T, Sakai T, Sugimoto M, et al. 1991; Factor IX Amagasaki: a new mutation in the catalytic domain resulting in the loss of both coagulant and esterase activities. Biochemistry. 30:11286–91. DOI: 10.1021/bi00111a014. PMID: 1958666.
Article27. Gao W, Xu Y, Liu H, et al. 2020; Characterization of missense mutations in the signal peptide and propeptide of FIX in hemophilia B by a cell-based assay. Blood Adv. 4:3659–67. DOI: 10.1182/bloodadvances.2020002520. PMID: 32766856. PMCID: PMC7422117.
Article28. Richards S, Aziz N, Bale S, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus re-commendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article29. Chavali S, Sharma A, Tabassum R, Bharadwaj D. 2008; Sequence and structural properties of identical mutations with varying phenotypes in human coagulation factor IX. Proteins. 73:63–71. DOI: 10.1002/prot.22035. PMID: 18393396.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of a shared F8 mutation in the Korean patients with acquired hemophilia A
- Sequence variation data of F8 and F9 genes in functionally validated control individuals: implications on the molecular diagnosis of hemophilia
- Direct detection of hemophilia B F9 gene mutation using multiplex PCR and conformation sensitive gel electrophoresis
- BRIP1/FANCJ Mutation Analysis in a Family with History of Male and Female Breast Cancer in India
- The First Indian Patient With Benign Hereditary Chorea due to a De Novo Mutation in the NKX2-1 Gene