J Breast Cancer.
2017 Mar;20(1):104-107. 10.4048/jbc.2017.20.1.104.
BRIP1/FANCJ Mutation Analysis in a Family with History of Male and Female Breast Cancer in India
- Affiliations
-
- 1Department of Oncologic Sciences, Mitchell Cancer Institute, University of South Alabama, Mobile, USA. kpalle@health.southalabama.edu
- 2Institute of Genetics and Hospital for Genetic Diseases, Osmania University, Hyderabad, India.
- 3Centre for Cellular and Molecular Biology, Hyderabad, India.
- 4Mehdi Nawaz Jung Institute of Oncology & Regional Centre, Hyderabad, India.
- KMID: 2379405
- DOI: http://doi.org/10.4048/jbc.2017.20.1.104
Abstract
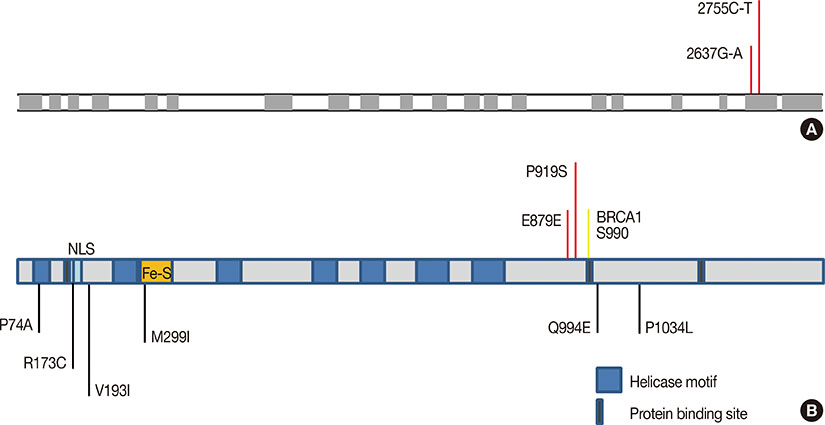
- Male breast cancer (MBC) is a rare and poorly studied disease that is a growing global health problem. Interestingly, both the molecular basis of MBC and its histological profile are often quite distinct from the far more prevalent female breast cancer, emphasizing the need for increased focus on MBC. Here, we present a case report of an MBC patient from India with a strong familial history of breast cancer. This patient was normal for BRCA1/2 and many other common breast cancer-associated genes. However, upon further analysis, the individual was found to possess two mutations in the DNA helicase and tumor suppressor gene BRIP1, including a silent mutation at residue 879 as well as a P919S variant. Other family members were also screened for these mutations. To the best of our knowledge, this is the first report of BRIP1 mutation in MBC in the Indian population.
MeSH Terms
Figure
Reference
-
1. Korde LA, Zujewski JA, Kamin L, Giordano S, Domchek S, Anderson WF, et al. Multidisciplinary meeting on male breast cancer: summary and research recommendations. J Clin Oncol. 2010; 28:2114–2122.
Article2. Weiss JR, Moysich KB, Swede H. Epidemiology of male breast cancer. Cancer Epidemiol Biomarkers Prev. 2005; 14:20–26.3. Kalyani R, Das S, Bindra Singh MS, Kumar H. Cancer profile in the Department of Pathology of Sri Devaraj Urs Medical College, Kolar: a ten years study. Indian J Cancer. 2010; 47:160–165.
Article4. Deb S, Jene N, Fox SB. Kconfab Investigators. Genotypic and phenotypic analysis of familial male breast cancer shows under representation of the HER2 and basal subtypes in BRCA-associated carcinomas. BMC Cancer. 2012; 12:510.
Article5. Yu E, Stitt L, Vujovic O, Joseph K, Assouline A, Younus J, et al. Male breast cancer prognostic factors versus female counterparts with propensity scores and matched-pair analysis. Cureus. 2015; 7:e355.
Article6. Evans DG, Bulman M, Young K, Howard E, Bayliss S, Wallace A, et al. BRCA1/2 mutation analysis in male breast cancer families from North West England. Fam Cancer. 2008; 7:113–117.
Article7. Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001; 105:149–160.
Article8. Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31:3812–3814.
Article9. National Cancer Institute. SEER cancer statistics review, 1975-2013. 2017. Accessed January 17th. http://seer.cancer.gov/csr/1975_2013/.10. Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006; 38:1239–1241.
Article11. Brosh RM Jr, Cantor SB. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front Genet. 2014; 5:372.
Article12. Clark DW, Tripathi K, Dorsman JC, Palle K. FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation. Oncotarget. 2015; 6:28816–28832.
Article13. Sigurdson AJ, Hauptmann M, Chatterjee N, Alexander BH, Doody MM, Rutter JL, et al. Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer. 2004; 4:9.
Article14. Vahteristo P, Yliannala K, Tamminen A, Eerola H, Blomqvist C, Nevanlinna H. BACH1 Ser919Pro variant and breast cancer risk. BMC Cancer. 2006; 6:19.
Article15. Silvestri V, Rizzolo P, Falchetti M, Zanna I, Masala G, Bianchi S, et al. Mutation analysis of BRIP1 in male breast cancer cases: a population-based study in Central Italy. Breast Cancer Res Treat. 2011; 126:539–543.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of BRIP1 Variants among Korean Patients with BRCA1/2 Mutation-Negative High-Risk Breast Cancer
- Interactions of Family History of Breast Cancer with Radiotherapy in Relation to the Risk of Breast Cancer Recurrence
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- Total Cholesterol and Triglyceride Levels in Patients with Breast Cancer
- Prostate Cancer in a Patient with a Family History of BRCA Mutation: a Case Report and Literature Review