Ann Surg Treat Res.
2021 Dec;101(6):322-331. 10.4174/astr.2021.101.6.322.
A novel strategy to promote liver regeneration: utilization of secretome obtained from survivin-overexpressing adipose-derived stem cells
- Affiliations
-
- 1Department of Surgery, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2Catholic Central Laboratory of Surgery, Institute of Biomedical Industry, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Department of Surgery, Eunpyeong St. Mary̓s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2522929
- DOI: http://doi.org/10.4174/astr.2021.101.6.322
Abstract
- Purpose
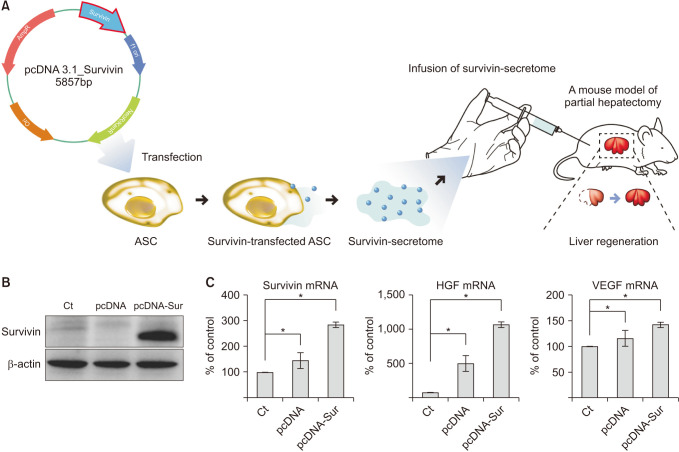
Survivin is a typical antiapoptotic protein. It is copiously expressed during human fetal development but is infrequently present in adult tissues. In this experiment, we researched the treatment effect of the secretome that adiposederived stem cells (ASCs) transfected with survivin.
Methods
First of all, we generated survivin-overexpressing ASCs transfected with a plasmid comprising a gene encoding survivin. The secreted substances released from survivin-overexpressing ASCs (survivin-secretome) were collected, and were determined their in vitro and in vivo therapeutic potential, especially in the model of liver impairment.
Results
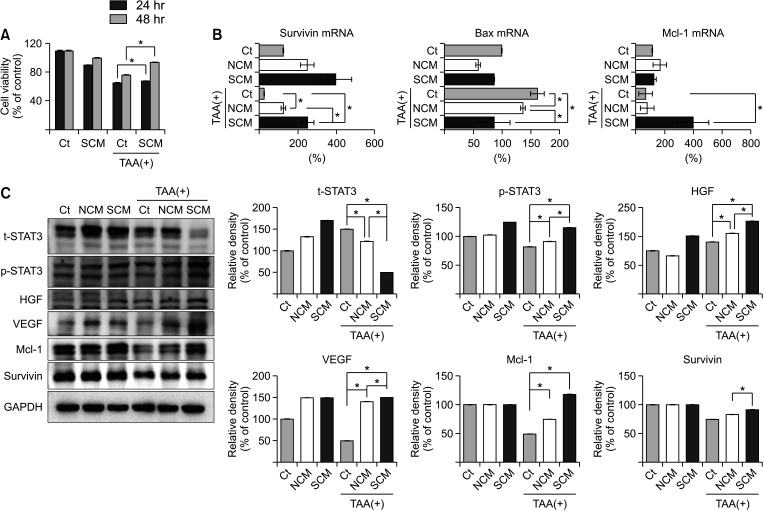
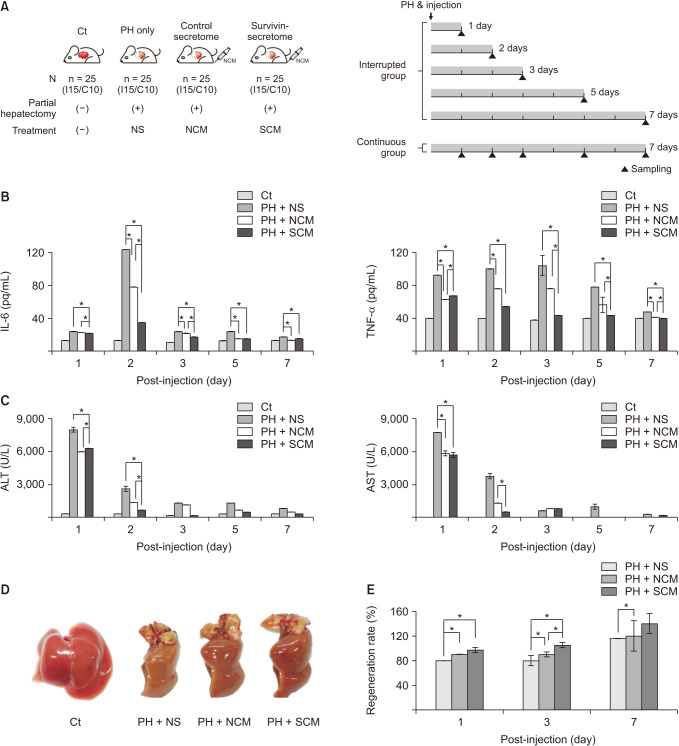
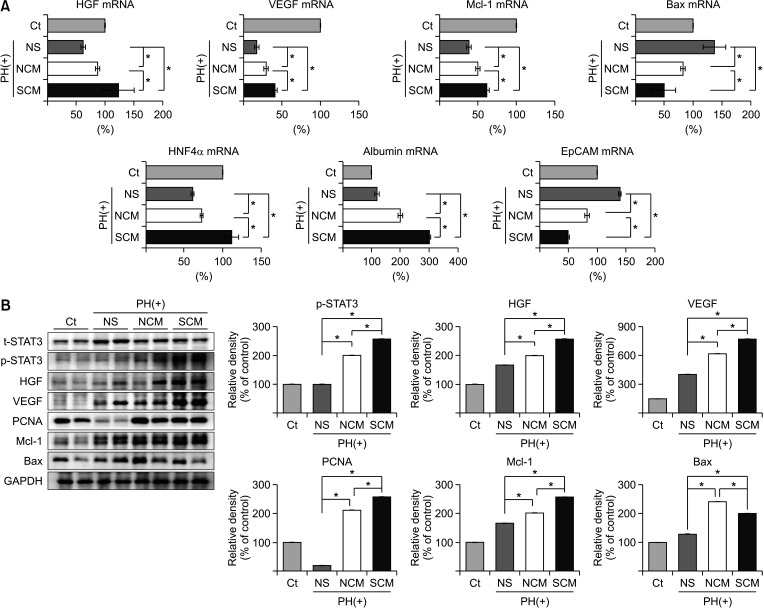
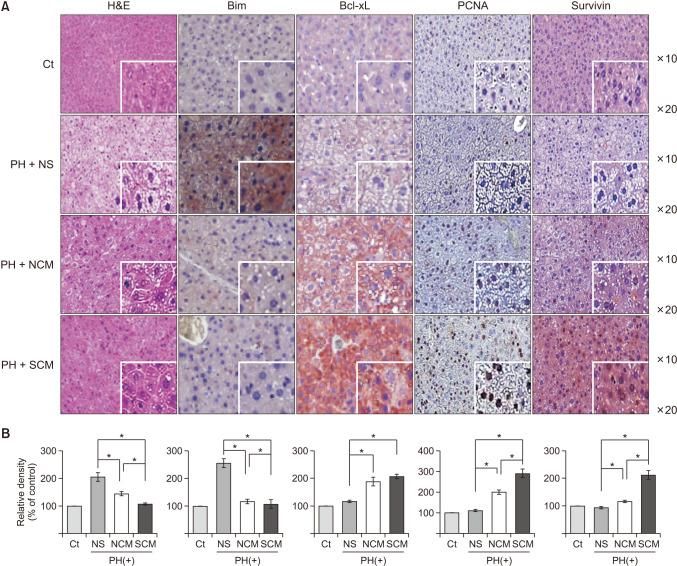
In vitro, the survivin-secretome significantly increased cell viability and promoted the expression of proliferationrelated markers (proliferating cell nuclear antigen [PCNA], phospho-signal transducer and activator of transcription 3 (p-STAT3), hepatocyte growth factor [HGF], vascular endothelial growth factor [VEGF]) and anti-apoptosis-related markers (myeloid cell leukemia-1 [Mcl-1] and survivin) (P < 0.05). In vivo using 70% hepatectomy mice, the survivin-secretome group exhibited the lowest serum levels of interleukin-6, tumor necrosis factor-α (P < 0.05). The serum levels of liver transaminases (alanine aminotransferase and aspartate aminotransferase) were also the lowest in the survivin-secretome group (P < 0.05). The survivin-secretome group also exhibited the highest liver regeneration on the 7th day after 70% partial hepatectomy (P < 0.05). In the subsequent liver specimen analysis, the specimens of survivin-secretome exhibited the highest expression of p-STAT3, HGF, VEGF, PCNA, and Mcl-1 and the lowest expression of bcl-2-like protein 4 (P < 0.05).
Conclusion
Taken together, secretome secreted by survivin-overexpressing ASCs could be an effective way to improve liver regeneration and repair for liver injury treatment.
Figure
Reference
-
1. Farid H, O’Connell T. Hepatic resections: changing mortality and morbidity. Am Surg. 1994; 60:748–752. PMID: 7944036.2. Liu J, Zhang Y, Zhu H, Qiu L, Guo C. Prediction of perioperative outcome after hepatic resection for pediatric patients. BMC Gastroenterol. 2019; 19:201. PMID: 31775648.
Article3. Itoh T, Miyajima A. Liver regeneration by stem/progenitor cells. Hepatology. 2014; 59:1617–1626. PMID: 24115180.
Article4. Puglisi MA, Saulnier N, Piscaglia AC, Tondi P, Agnes S, Gasbarrini A. Adipose tissue-derived mesenchymal stem cells and hepatic differentiation: old concepts and future perspectives. Eur Rev Med Pharmacol Sci. 2011; 15:355–364. PMID: 21608430.5. Baglio SR, Pegtel DM, Baldini N. Mesenchymal stem cell secreted vesicles provide novel opportunities in (stem) cell-free therapy. Front Physiol. 2012; 3:359. PMID: 22973239.
Article6. Filip S, Mokrý J, English D, Vojácek J. Stem cell plasticity and issues of stem cell therapy. Folia Biol (Praha). 2005; 51:180–187. PMID: 16419613.7. Makridakis M, Roubelakis MG, Vlahou A. Stem cells: insights into the secretome. Biochim Biophys Acta. 2013; 1834:2380–2384. PMID: 23376432.
Article8. Kim KH, Lee JI, Kim OH, Hong HE, Kwak BJ, Choi HJ, et al. Ameliorating liver fibrosis in an animal model using the secretome released from miR-122-transfected adipose-derived stem cells. World J Stem Cells. 2019; 11:990–1004. PMID: 31768225.
Article9. Lee J, Choi JH, Joo CK. TGF-β1 regulates cell fate during epithelial-mesenchymal transition by upregulating survivin. Cell Death Dis. 2013; 4:e714. PMID: 23828577.
Article10. Deguchi M, Shiraki K, Inoue H, Okano H, Ito T, Yamanaka T, et al. Expression of survivin during liver regeneration. Biochem Biophys Res Commun. 2002; 297:59–64. PMID: 12220508.
Article11. Hagemann S, Wohlschlaeger J, Bertram S, Levkau B, Musacchio A, Conway EM, et al. Loss of Survivin influences liver regeneration and is associated with impaired Aurora B function. Cell Death Differ. 2013; 20:834–844. PMID: 23519077.
Article12. Park JH, Kim OH, Kim KH, Hong HE, Seo H, Choi HJ, et al. Isolation of secretome with enhanced antifibrotic properties from miR-214-transfected adipose-derived stem cells. J Korean Med Sci. 2019; 34:e273. PMID: 31760709.
Article13. Greene AK, Puder M. Partial hepatectomy in the mouse: technique and perioperative management. J Invest Surg. 2003; 16:99–102. PMID: 12746193.
Article14. Park MR, Wong MS, Araúzo-Bravo MJ, Lee H, Nam D, Park SY, et al. Oct4 and Hnf4α-induced hepatic stem cells ameliorate chronic liver injury in liver fibrosis model. PLoS One. 2019; 14:e0221085. PMID: 31404112.
Article15. Zhao Q, Zhou H, Liu Q, Cao Y, Wang G, Hu A, et al. Prognostic value of the expression of cancer stem cell-related markers CD133 and CD44 in hepatocellular carcinoma: from patients to patient-derived tumor xenograft models. Oncotarget. 2016; 7:47431–47443. PMID: 27329727.
Article16. Lee TH, Park DS, Jang JY, Lee I, Kim JM, Choi GS, et al. Human placenta hydrolysate promotes liver regeneration via activation of the cytokine/growth factor-mediated pathway and anti-oxidative effect. Biol Pharm Bull. 2019; 42:607–616. PMID: 30930420.17. Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003; 22:8581–8589. PMID: 14634620.
Article18. O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A. 2000; 97:13103–13107. PMID: 11069302.19. Ebrahimiyan H, Aslani S, Rezaei N, Jamshidi A, Mahmoudi M. Survivin and autoimmunity; the ins and outs. Immunol Lett. 2018; 193:14–24. PMID: 29155234.
Article20. Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000; 31:1080–1085. PMID: 10796883.
Article21. Deveraux QL, Reed JC. IAP family proteins: suppressors of apoptosis. Genes Dev. 1999; 13:239–252. PMID: 9990849.22. Sun C, Cai M, Gunasekera AH, Meadows RP, Wang H, Chen J, et al. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature. 1999; 401:818–822. PMID: 10548111.
Article23. Shi Y. Survivin structure: crystal unclear. Nat Struct Biol. 2000; 7:620–623. PMID: 10932241.24. Conway EM, Pollefeyt S, Steiner-Mosonyi M, Luo W, Devriese A, Lupu F, et al. Deficiency of survivin in transgenic mice exacerbates Fas-induced apoptosis via mitochondrial pathways. Gastroenterology. 2002; 123:619–631. PMID: 12145814.
Article25. Suzuki A, Hayashida M, Ito T, Kawano H, Nakano T, Miura M, et al. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000; 19:3225–3234. PMID: 10918579.
Article26. Wang P, Zhen H, Zhang J, Zhang W, Zhang R, Cheng X, et al. Survivin promotes glioma angiogenesis through vascular endothelial growth factor and basic fibroblast growth factor in vitro and in vivo. Mol Carcinog. 2012; 51:586–595. PMID: 21761458.
Article27. Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, et al. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood. 2011; 117:352–361. PMID: 20930069.
Article28. Tran J, Master Z, Yu JL, Rak J, Dumont DJ, Kerbel RS. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc Natl Acad Sci U S A. 2002; 99:4349–4354. PMID: 11917134.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Liver Regenerating Potential of the Secretome Obtained from Adipose-derived Stem Cells Cultured under the Hypoxic Environment
- Adipose Tissue - Adequate, Accessible Regenerative Material
- Isolation of Secretome with Enhanced Antifibrotic Properties from miR-214-Transfected Adipose-Derived Stem Cells
- Bone Regeneration from Adipose Stem Cells
- Pioneering PGC-1αα–boosted secretome: a novel approach to combating liver fibrosis