Int J Stem Cells.
2021 Nov;14(4):423-433. 10.15283/ijsc21080.
miR-200c-141 Enhances Sheep Kidney Cell Reprogramming into Pluripotent Cells by Targeting ZEB1
- Affiliations
-
- 1College of Animal Science and Technology, Shihezi University, Xinjiang, China
- 2Collaborative Innovation Center for Prevention and Control of High Incidence Zoonotic Infectious, Shihezi, China
- 3State Key Laboratory of Sheep Genetic Improvement and Healthy Production/Xinjiang Academy of Agricultural and Reclamation Sciences, Xinjiang, China
- 4College of Life Technology, Shihezi University, Xinjiang, China
- KMID: 2522561
- DOI: http://doi.org/10.15283/ijsc21080
Abstract
- Background and Objectives
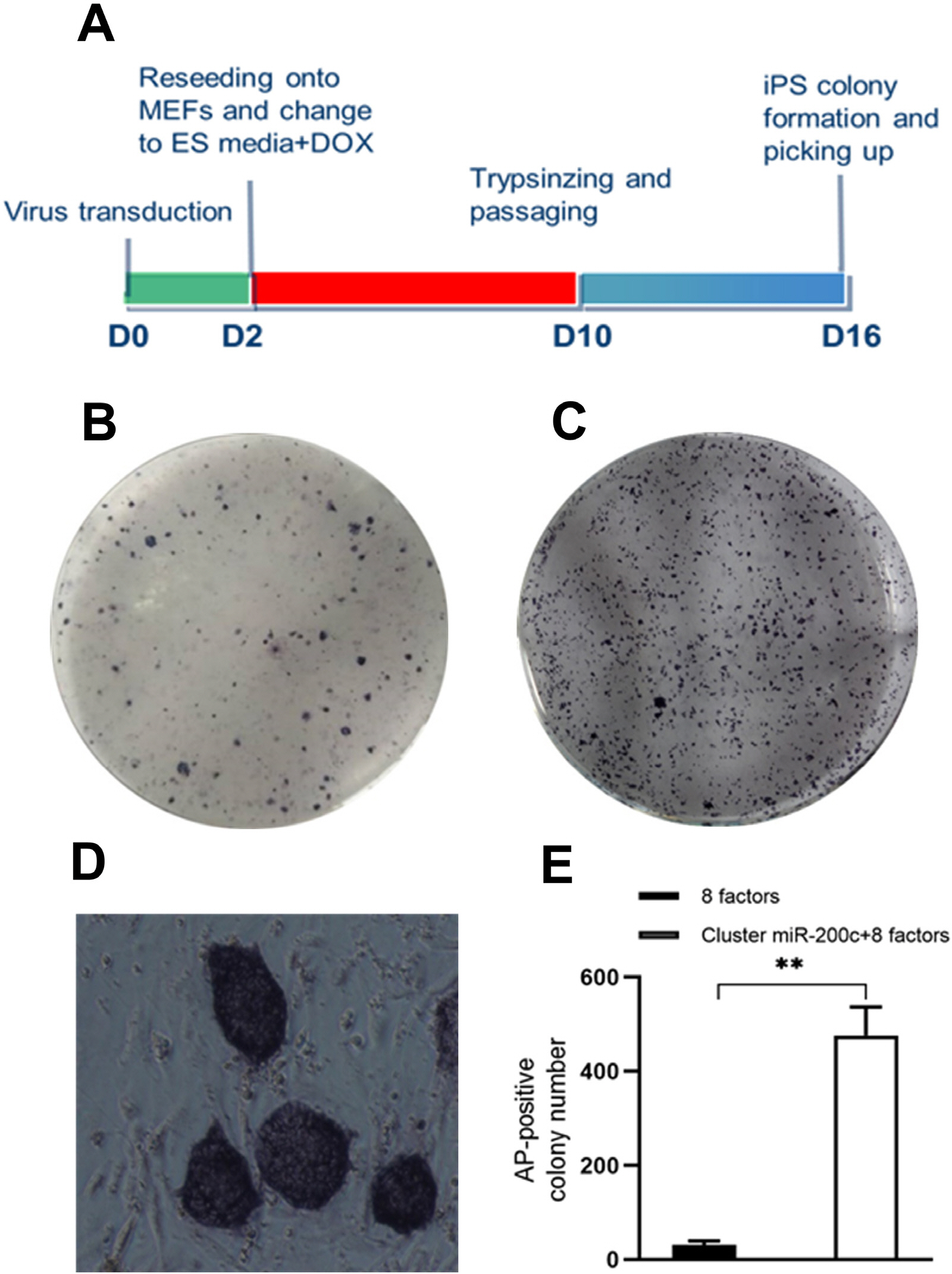
Sheep-induced pluripotent stem cells (siPSCs) have low reprogramming efficiency, thereby hampering their use in biotechnology and agriculture. Several studies have shown that some microRNAs play an important role in promoting somatic reprogramming in mouse and human. In this study, we investigated the effect of miR-200c-141 on somatic reprogramming in sheep and explored the mechanism of promoting the reprogramming.
Methods and Results
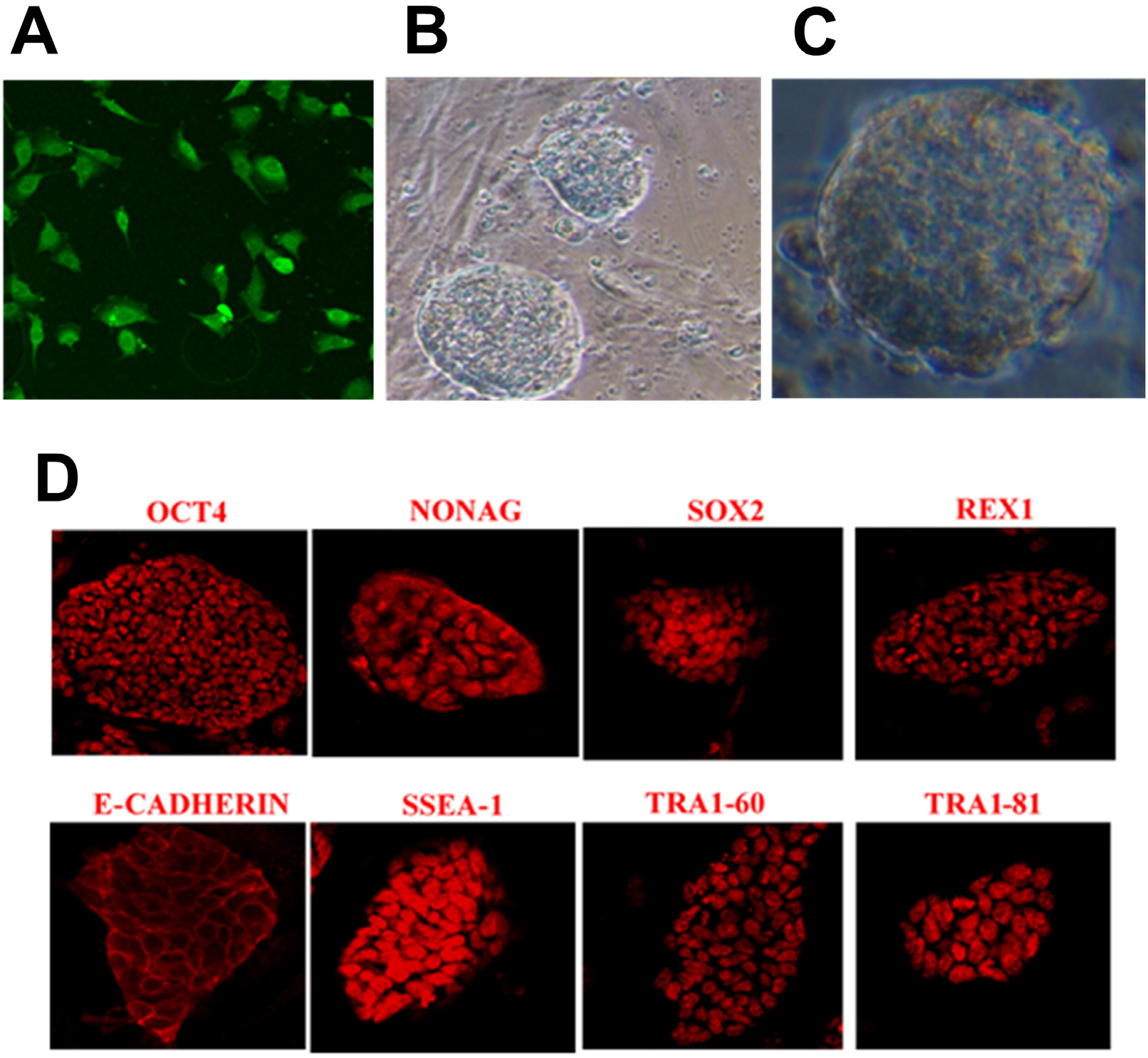
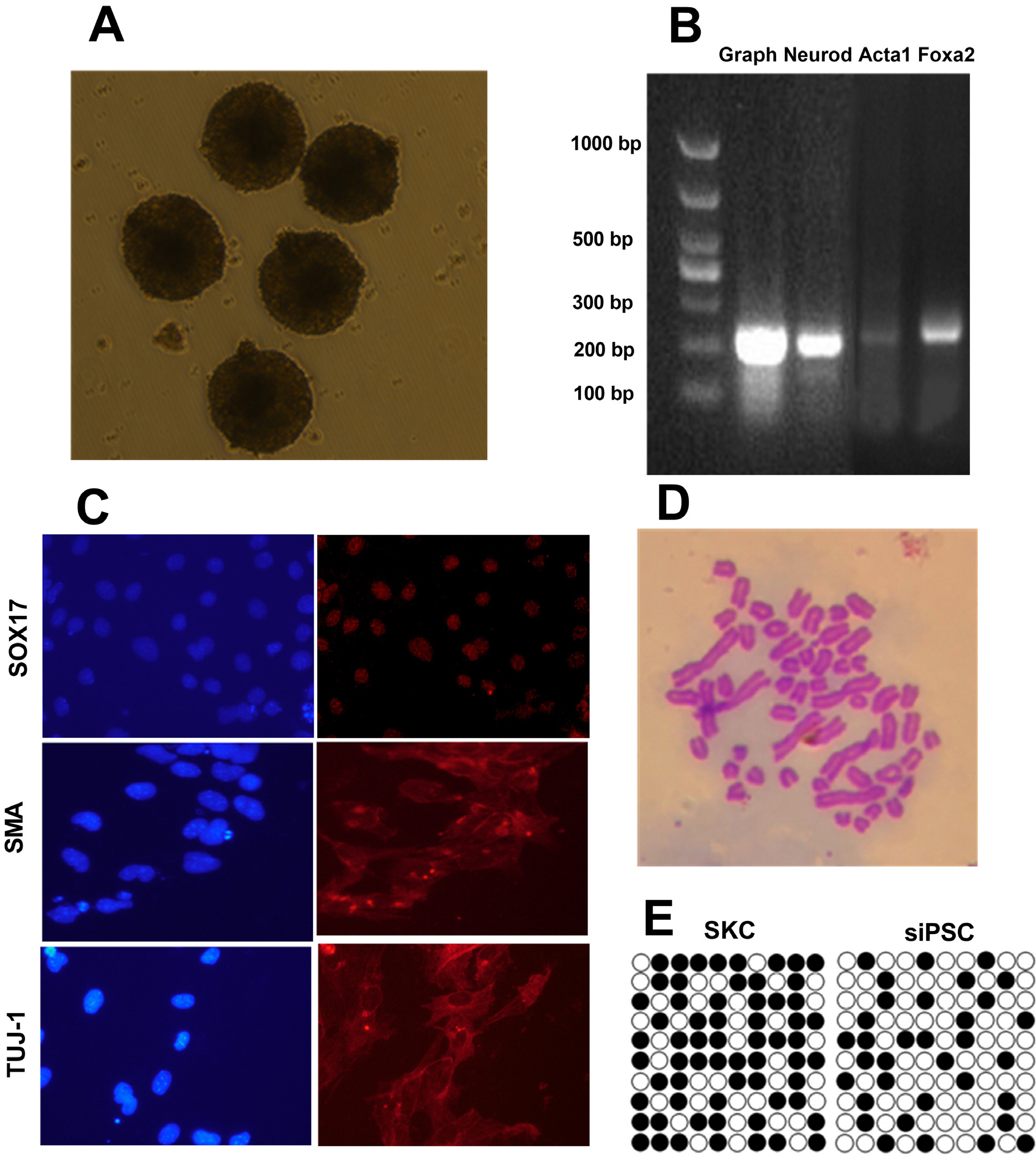
The lentivirus system driven by tetracycline (TET)-on carrying Oct4, Sox2, c-Myc, Klf4, Nanog, Lin28, hTERT, and SV40LT (OSKMNLST) could reprogram sheep kidney cells into pluripotent cells. Overexpression of miR-200c-141 in combination with OSKMNLST could significantly improve the efficiency of sheep iPSC generation (p<0.01). Sheep iPSCs derived from miR-200c-141 showed embryonic stem cell (ESC)-like pluripotent properties, were positive for alkaline phosphatase and some pluripotent markers by quantitative real-time PCR (qRT-PCR) and immunofluorescence, and were able to differentiate into three germ layers in vitro. Oar-miR-200c was transfected into HEK293FT cells and was able to target the zinc finger E-box-binding homeobox 1 (ZEB1) 3’UTR using dual luciferase reporting analysis. Overexpression of oar-miR-200c in SKCs significantly reduced the expression of ZEB1, but increased the expression of E-cadherin by qRT-PCR and western blotting analysis.
Conclusions
These results suggest that miR-200c-141 can promote the reprogramming of sheep somatic cells to iPSCs, and oar-miR-200c targeted ZEB1 3’UTR, significantly decreased expression of ZEB1, and increased expression of E-cadherin. Oar-miR-200c may improve the MET process by affecting the TGF-β signaling pathway, thus improving the efficiency of somatic cell reprogramming in sheep.
Keyword
- Sheep; iPSC; miR-200c; ZEB1; E-cadherin
Figure
Reference
-
References
1. Takahashi K, Yamanaka S. 2006; Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676. DOI: 10.1016/j.cell.2006.07.024. PMID: 16904174.
Article2. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. 2007; Induced pluripotent stem cell lines derived from human somatic cells. Science. 318:1917–1920. DOI: 10.1126/science.1151526. PMID: 18029452.
Article3. Li Y, Cang M, Lee AS, Zhang K, Liu D. 2011; Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS One. 6:e15947. DOI: 10.1371/journal.pone.0015947. PMID: 21253598. PMCID: PMC3017083.
Article4. Bao L, He L, Chen J, Wu Z, Liao J, Rao L, Ren J, Li H, Zhu H, Qian L, Gu Y, Dai H, Xu X, Zhou J, Wang W, Cui C, Xiao L. 2011; Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 21:600–608. DOI: 10.1038/cr.2011.6. PMID: 21221129. PMCID: PMC3203662.
Article5. Liu J, Balehosur D, Murray B, Kelly JM, Sumer H, Verma PJ. 2012; Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology. 77:338–346.e1. DOI: 10.1016/j.theriogenology.2011.08.006. PMID: 21958637.
Article6. Shi H, Fu Q, Li G, Ren Y, Hu S, Ni W, Guo F, Shi M, Meng L, Zhang H, Qiao J, Guo Z, Chen C. 2015; Roles of p53 and ASF1A in the reprogramming of sheep kidney cells to pluripotent cells. Cell Reprogram. 17:441–452. DOI: 10.1089/cell.2015.0039. PMID: 26580119. PMCID: PMC4677545.
Article7. Sartori C, DiDomenico AI, Thomson AJ, Milne E, Lillico SG, Burdon TG, Whitelaw CB. 2012; Ovine-induced pluripotent stem cells can contribute to chimeric lambs. Cell Reprogram. 14:8–19. DOI: 10.1089/cell.2011.0050. PMID: 22217199.
Article8. Su Y, Zhu J, Salman S, Tang Y. 2020; Induced pluripotent stem cells from farm animals. J Anim Sci. 98:skaa343. DOI: 10.1093/jas/skaa343. PMID: 33098420. PMCID: PMC7660146.
Article9. Pfaff N, Moritz T, Thum T, Cantz T. 2012; miRNAs involved in the generation, maintenance, and differentiation of pluripotent cells. J Mol Med (Berl). 90:747–752. DOI: 10.1007/s00109-012-0922-z. PMID: 22684238.
Article10. Choi E, Choi E, Hwang KC. 2013; MicroRNAs as novel regulators of stem cell fate. World J Stem Cells. 5:172–187. DOI: 10.4252/wjsc.v5.i4.172. PMID: 24179605. PMCID: PMC3812521.
Article11. Houbaviy HB, Murray MF, Sharp PA. 2003; Embryonic stem cell-specific MicroRNAs. Dev Cell. 5:351–358. DOI: 10.1016/S1534-5807(03)00227-2. PMID: 12919684.
Article12. Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS. 2004; Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 270:488–498. DOI: 10.1016/j.ydbio.2004.02.019. PMID: 15183728.
Article13. Judson RL, Babiarz JE, Venere M, Blelloch R. 2009; Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 27:459–461. DOI: 10.1038/nbt.1535. PMID: 19363475. PMCID: PMC2743930.
Article14. Li N, Long B, Han W, Yuan S, Wang K. 2017; microRNAs: important regulators of stem cells. Stem Cell Res Ther. 8:110. DOI: 10.1186/s13287-017-0551-0. PMID: 28494789. PMCID: PMC5426004.
Article15. Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. 2011; Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 8:376–388. DOI: 10.1016/j.stem.2011.03.001. PMID: 21474102. PMCID: PMC3090650.
Article16. Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, Gao L, Ye D, Zhou Y, Chen J, Wang J, Wu M, Liu H, Kang J. 2013; Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci U S A. 110:2858–2863. DOI: 10.1073/pnas.1212769110. PMID: 23386720. PMCID: PMC3581874.
Article17. Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. 2011; Repro-gramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 8:633–638. DOI: 10.1016/j.stem.2011.05.001. PMID: 21620789.
Article18. Mallanna SK, Rizzino A. 2010; Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 344:16–25. DOI: 10.1016/j.ydbio.2010.05.014. PMID: 20478297. PMCID: PMC2935203.
Article19. Livak KJ, Schmittgen TD. 2001; Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. DOI: 10.1006/meth.2001.1262. PMID: 11846609.
Article20. Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, Qin B, Zhang R, Wu J, Lai L, Teng M, Niu L, Zhang B, Esteban MA, Pei D. 2011; MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 286:17359–17364. DOI: 10.1074/jbc.C111.235960. PMID: 21454525. PMCID: PMC3089577.
Article21. Du X, Feng T, Yu D, Wu Y, Zou H, Ma S, Feng C, Huang Y, Ouyang H, Hu X, Pan D, Li N, Wu S. 2015; Barriers for deriving transgene-free pig iPS cells with episomal vectors. Stem Cells. 33:3228–3238. DOI: 10.1002/stem.2089. PMID: 26138940. PMCID: PMC5025037.
Article22. Ma K, Song G, An X, Fan A, Tan W, Tang B, Zhang X, Li Z. 2014; miRNAs promote generation of porcine-induced pluripotent stem cells. Mol Cell Biochem. 389:209–218. DOI: 10.1007/s11010-013-1942-x. PMID: 24464032.
Article23. Pillai VV, Kei TG, Reddy SE, Das M, Abratte C, Cheong SH, Selvaraj V. 2019; Induced pluripotent stem cell generation from bovine somatic cells indicates unmet needs for pluripotency sustenance. Anim Sci J. 90:1149–1160. DOI: 10.1111/asj.13272. PMID: 31322312.
Article24. Tai D, Liu P, Gao J, Jin M, Xu T, Zuo Y, Liang H, Liu D. 2015; Generation of Arbas cashmere goat induced pluripotent stem cells through fibroblast reprogramming. Cell Reprogram. 17:297–305. DOI: 10.1089/cell.2014.0107. PMID: 26731591.
Article25. Sandmaier SE, Nandal A, Powell A, Garrett W, Blomberg L, Donovan DM, Talbot N, Telugu BP. 2015; Generation of induced pluripotent stem cells from domestic goats. Mol Reprod Dev. 82:709–721. DOI: 10.1002/mrd.22512. PMID: 26118622.
Article26. Bobik A. 2006; Transforming growth factor-betas and vascular disorders. Arterioscler Thromb Vasc Biol. 26:1712–1720. DOI: 10.1161/01.ATV.0000225287.20034.2c. PMID: 16675726.27. Moreno-Bueno G, Cubillo E, Sarrió D, Peinado H, Rodríguez-Pinilla SM, Villa S, Bolós V, Jordá M, Fabra A, Portillo F, Palacios J, Cano A. 2006; Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res. 66:9543–9556. DOI: 10.1158/0008-5472.CAN-06-0479. PMID: 17018611.
Article28. Shirakihara T, Saitoh M, Miyazono K. 2007; Differential regulation of epithelial and mesenchymal markers by deltaEF1 +proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 18:3533–3544. DOI: 10.1091/mbc.e07-03-0249. PMID: 17615296. PMCID: PMC1951739.
Article29. Peinado H, Olmeda D, Cano A. 2007; Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 7:415–428. DOI: 10.1038/nrc2131. PMID: 17508028.
Article30. Bryant JL, Britson J, Balko JM, Willian M, Timmons R, Frolov A, Black EP. 2012; A microRNA gene expression signature predicts response to erlotinib in epithelial cancer cell lines and targets EMT. Br J Cancer. 106:148–156. DOI: 10.1038/bjc.2011.465. PMID: 22045191. PMCID: PMC3251842.
Article31. Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. 2008; A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9:582–589. DOI: 10.1038/embor.2008.74. PMID: 18483486. PMCID: PMC2396950.
Article32. Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. 2008; A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68:7846–7854. DOI: 10.1158/0008-5472.CAN-08-1942. PMID: 18829540.
Article33. Brabletz S, Brabletz T. 2010; The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 11:670–677. DOI: 10.1038/embor.2010.117. PMID: 20706219. PMCID: PMC2933868.
Article34. Lamouille S, Xu J, Derynck R. 2014; Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 15:178–196. DOI: 10.1038/nrm3758. PMID: 24556840. PMCID: PMC4240281.
Article35. Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. 2009; A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 5:491–503. DOI: 10.1016/j.stem.2009.09.012. PMID: 19818703. PMCID: PMC3335195.
Article36. Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, Ding S. 2009; A chemical platform for improved induction of human iPSCs. Nat Methods. 6:805–808. DOI: 10.1038/nmeth.1393. PMID: 19838168. PMCID: PMC3724527.
Article37. Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. 2010; Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 7:64–77. DOI: 10.1016/j.stem.2010.04.015. PMID: 20621051.
Article38. Peter ME. 2009; Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 8:843–852. DOI: 10.4161/cc.8.6.7907. PMID: 19221491. PMCID: PMC2688687.
Article39. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008; The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601. DOI: 10.1038/ncb1722. PMID: 18376396.
Article40. Korpal M, Lee ES, Hu G, Kang Y. 2008; The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 283:14910–14914. DOI: 10.1074/jbc.C800074200. PMID: 18411277. PMCID: PMC3258899.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Inhibition of Class I Histone Deacetylase Enhances Self-Reprogramming of Spermatogonial Stem Cells into Pluripotent Stem Cells
- miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple Negative Breast Cancer
- Lineage-specific Expression of miR-200 Family in Human Embryonic Stem Cells during In Vitro Differentiation
- MicroRNA-200c as a Prognostic Biomarker for Pancreatic Cancer
- Reprogramming of Cancer Cells into Induced Pluripotent Stem Cells Questioned