J Pathol Transl Med.
2020 Nov;54(6):453-461. 10.4132/jptm.2020.08.13.
Liquid biopsy using extracellular vesicle–derived DNA in lung adenocarcinoma
- Affiliations
-
- 1Precision Medicine Lung Cancer Center, Konkuk University Medical Center, Seoul, Korea
- 2Department of Pulmonary Medicine, Konkuk University School of Medicine, Seoul, Korea
- 3Department of Pathology, Konkuk University School of Medicine, Seoul, Korea
- KMID: 2509498
- DOI: http://doi.org/10.4132/jptm.2020.08.13
Abstract
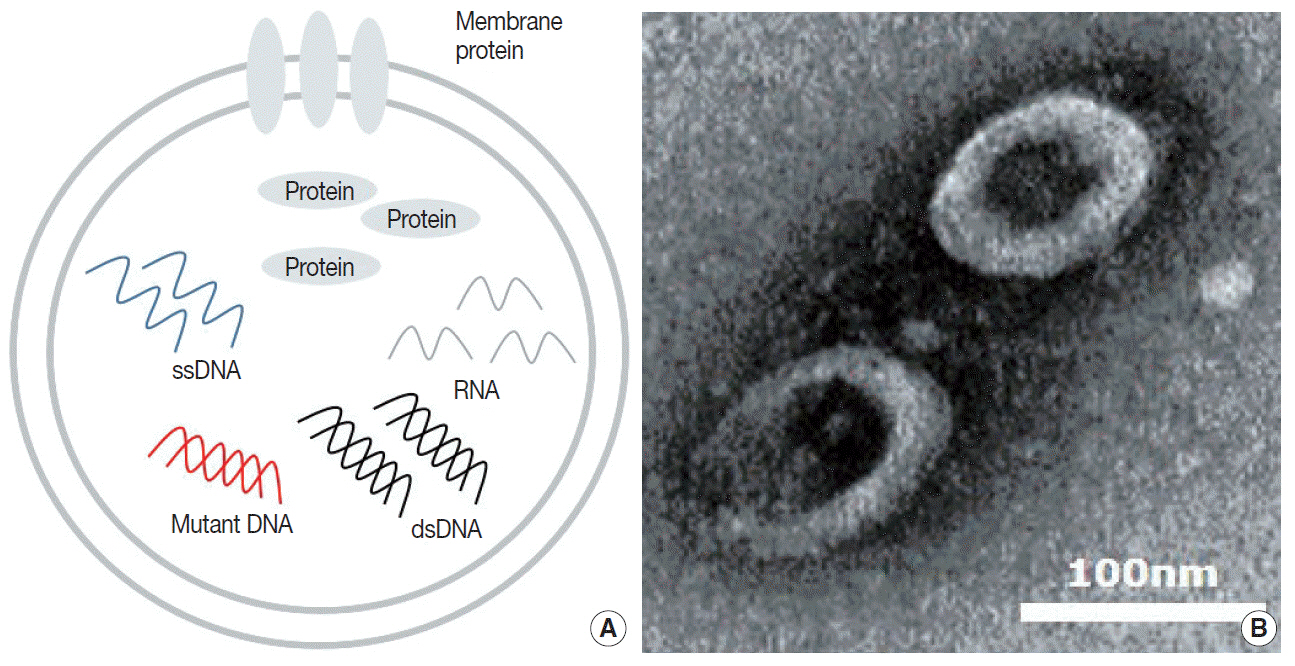
- Blood liquid biopsy has emerged as a way of overcoming the clinical limitations of repeat biopsy by testing for the presence of acquired resistance mutations to therapeutic agents. Despite its merits of repeatability and non-invasiveness, this method is currently only used as a supplemental test due to a relatively low sensitivity rate of 50%–60%, and cannot replace tissue biopsy. The circulating tumor DNAs used in blood liquid biopsies are passive products of fragmented DNA with a short half-life released following tumor cell death; the low sensitivity seen with liquid blood biopsy results from this instability, which makes increasing the sensitivity of this test fundamentally difficult. Extracellular vesicles (EVs) are ideal carriers of cancer biomarkers, as cancer cells secret an abundance of EVs, and the contents of tumor cell-originated EVs reflect the molecular and genetic composition of parental cells. In addition, EV-derived DNAs (EV DNAs) consist of large-sized genomic DNAs and tumor-specific oncogenic mutant DNAs. For these reasons, liquid biopsy using EV DNA has the potential to overcome issues arising from tissue shortages associated with small biopsies, which are often seen in lung cancer patients, and the biopsy product can be used in other diagnostic methods, such as epidermal growth factor receptor (EGFR) mutation testing and next-generation sequencing (NGS). A higher sensitivity can be achieved when EV DNAs obtained from bronchoalveolar lavage fluid (BALF) are used rather than those from blood. BALF, when obtained close to the tumor site, is a promising liquid biopsy tool, as it enables the gathering of both cellular and non-cellular fractions of the tumor microenvironment, and provides increased diagnostic sensitivity when compared to blood.
Keyword
Figure
Reference
-
References
1. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019; 85:8.
Article2. de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018; 7:220–33.
Article3. Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011; 32:605–44.
Article4. Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018; 17:38.
Article5. Imakita T, Matsumoto H, Hirano K, Morisawa T, Sakurai A, Kataoka Y. Impact on prognosis of rebiopsy in advanced non-small cell lung cancer patients after epidermal growth factor receptor-tyrosine kinase inhibitor treatment: a systematic review. BMC Cancer. 2019; 19:105.
Article6. Bollinger MK, Agnew AS, Mascara GP. Osimertinib: A third-generation tyrosine kinase inhibitor for treatment of epidermal growth factor receptor-mutated non-small cell lung cancer with the acquired Thr790Met mutation. J Oncol Pharm Pract. 2018; 24:379–88.
Article7. Akamatsu H, Katakami N, Okamoto I, et al. Osimertinib in Japanese patients with EGFR T790M mutation-positive advanced non-small-cell lung cancer: AURA3 trial. Cancer Sci. 2018; 109:1930–8.
Article8. Odogwu L, Mathieu L, Goldberg KB, et al. FDA benefit-risk assessment of osimertinib for the treatment of metastatic non-small cell lung cancer harboring epidermal growth factor receptor T790M mutation. Oncologist. 2018; 23:353–9.
Article9. Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016; 7:48832–41.
Article10. Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018; 10:eaat4921.
Article11. Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJ. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018; 18:1174–96.
Article12. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014; 30:255–89.
Article13. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002; 2:569–79.
Article14. Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014; 289:3869–75.
Article15. Armstrong D, Wildman DE. Extracellular vesicles and the promise of continuous liquid biopsies. J Pathol Transl Med. 2018; 52:1–8.
Article16. Hur JY, Kim HJ, Lee JS, et al. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol Cancer. 2018; 17:15.
Article17. Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018; 116:46–54.
Article18. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013; 10:472–84.
Article19. Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease: latest advances and implications for cure. Nat Rev Clin Oncol. 2019; 16:409–24.20. Bracht JW, Mayo-de-Las-Casas C, Berenguer J, Karachaliou N, Rosell R. The present and future of liquid biopsies in non-small cell lung cancer: combining four biosources for diagnosis, prognosis, prediction, and disease monitoring. Curr Oncol Rep. 2018; 20:70.
Article21. Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta. 2014; 1846:539–46.
Article22. Calabuig-Farinas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. 2016; 5:466–82.23. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014; 4:650–61.
Article24. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004; 351:781–91.
Article25. Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012; 7:306–15.
Article26. Poudineh M, Sargent EH, Pantel K, Kelley SO. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat Biomed Eng. 2018; 2:72–84.
Article27. Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007; 450:1235–9.
Article28. Qian W, Zhang Y, Chen W. Capturing cancer: emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small. 2015; 11:3850–72.
Article29. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014; 32:579–86.
Article30. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977; 37:646–50.31. Karachaliou N, Sosa AE, Molina MA, Centelles Ruiz M, Rosell R. Possible application of circulating free tumor DNA in non-small cell lung cancer patients. J Thorac Dis. 2017; 9(Suppl 13):S1364–72.
Article32. Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019; 13:34.
Article33. Goldman JW, Noor ZS, Remon J, Besse B, Rosenfeld N. Are liquid biopsies a surrogate for tissue EGFR testing? Ann Oncol. 2018; 29:i38–46.
Article34. Hochmair MJ, Buder A, Schwab S, et al. Liquid-biopsy-based identification of EGFR T790M mutation-mediated resistance to afatinib treatment in patients with advanced EGFR mutation-positive NSCLC, and subsequent response to osimertinib. Target Oncol. 2019; 14:75–83.35. Rolfo C, Mack PC, Scagliotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018; 13:1248–68.36. Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.
Article37. Best MG, Wesseling P, Wurdinger T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018; 78:3407–12.
Article38. Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget. 2016; 7:1066–75.
Article39. Liu L, Lin F, Ma X, Chen Z, Yu J. Tumor-educated platelet as liquid biopsy in lung cancer patients. Crit Rev Oncol Hematol. 2020; 146:102863.
Article40. McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014; 16:717–27.
Article41. Best MG, Sol N, Kooi I, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015; 28:666–76.
Article42. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015; 34:474–90.
Article43. Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011; 9:86.
Article44. Carnino JM, Lee H, Jin Y. Isolation and characterization of extracellular vesicles from broncho-alveolar lavage fluid: a review and comparison of different methods. Respir Res. 2019; 20:240.
Article45. Aldick T, Bielaszewska M, Uhlin BE, Humpf HU, Wai SN, Karch H. Vesicular stabilization and activity augmentation of enterohaemorrhagic Escherichia coli haemolysin. Mol Microbiol. 2009; 71:1496–508.46. Dutta S, Iida K, Takade A, Meno Y, Nair GB, Yoshida S. Release of Shiga toxin by membrane vesicles in Shigella dysenteriae serotype 1 strains and in vitro effects of antimicrobials on toxin production and release. Microbiol Immunol. 2004; 48:965–9.47. Choi D, Lee TH, Spinelli C, Chennakrishnaiah S, D'Asti E, Rak J. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin Cell Dev Biol. 2017; 67:11–22.
Article48. Soldevilla B, Rodriguez M, San Millan C, et al. Tumor-derived exosomes are enriched in DeltaNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014; 23:467–78.49. Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012; 18:883–91.50. Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013; 32:2747–55.
Article51. Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014; 32:983–97.
Article52. Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014; 26:707–21.
Article53. Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015; 40:72–81.
Article54. Hsu YL, Hung JY, Chang WA, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017; 36:4929–42.
Article55. Li X, Wang S, Zhu R, Li H, Han Q, Zhao RC. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFkappaB-TLR signaling pathway. J Hematol Oncol. 2016; 9:42.
Article56. Fujita Y, Kosaka N, Araya J, Kuwano K, Ochiya T. Extracellular vesicles in lung microenvironment and pathogenesis. Trends Mol Med. 2015; 21:533–42.
Article57. Jung JH, Lee MY, Choi DY, et al. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics. 2015; 15:824–35.
Article58. Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009; 4:e5219.
Article59. Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015; 527:329–35.60. Lu Q, Zhang J, Allison R, et al. Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate. 2009; 69:411–8.61. Moon PG, Lee JE, Cho YE, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. 2016; 22:1757–66.
Article62. Rontogianni S, Synadaki E, Li B, et al. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019; 2:325.
Article63. Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017; 8:13048–58.
Article64. Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017; 21:2650–8.65. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014; 24:766–9.
Article66. Vagner T, Spinelli C, Minciacchi VR, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018; 7:1505403.
Article67. Kahlert C. Liquid biopsy: is there an advantage to analyzing circulating exosomal DNA compared to cfDNA or are they the same? Cancer Res. 2019; 79:2462–5.
Article68. Lazaro-Ibanez E, Lasser C, Shelke GV, et al. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles. 2019; 8:1656993.69. Sharma A, Johnson A. Exosome DNA: critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol. 2020; 235:1921–32.
Article70. Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011; 6:e23418.
Article71. Zill OA, Greene C, Sebisanovic D, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015; 5:1040–8.
Article72. San Lucas FA, Allenson K, Bernard V, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2016; 27:635–41.
Article73. Song Z, Cai Z, Yan J, Shao YW, Zhang Y. Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma. Transl Lung Cancer Res. 2019; 8:392–400.
Article74. Lee DH, Yoon H, Park S, et al. Urinary Exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci Rep. 2018; 8:14707.
Article75. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011; 11:426–37.
Article76. Hur JY, Lee JS, Kim IA, Kim HJ, Kim WS, Lee KY. Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl Lung Cancer Res. 2019; 8:1051–60.77. Bernard V, Kim DU, San Lucas FA, et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology. 2019; 156:108–18.
Article78. Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017; 28:741–7.79. Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686). Clin Cancer Res. 2016; 22:2386–95.80. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016; 34:3375–82.
Article81. Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016; 2:1014–22.82. Helman E, Nguyen M, Karlovich CA, et al. Cell-free DNA next-generation sequencing prediction of response and resistance to third-generation EGFR inhibitor. Clin Lung Cancer. 2018; 19:518–30.83. Brinkman K, Meyer L, Bickel A, et al. Extracellular vesicles from plasma have higher tumour RNA fraction than platelets. J Extracell Vesicles. 2020; 9:1741176.
Article84. Qi Y, Zha W, Zhang W. Exosomal miR-660-5p promotes tumor growth and metastasis in non-small cell lung cancer. J BUON. 2019; 24:599–607.85. Zhang Y, Zhang Y, Yin Y, Li S. Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol Res Pract. 2019; 215:152466.
Article86. Grimolizzi F, Monaco F, Leoni F, et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep. 2017; 7:15277.
Article87. Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017; 23:5311–9.
Article88. Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010; 28:1721–6.
Article89. Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018; 29:700–6.90. Morgensztern D, Waqar S, Subramanian J, Trinkaus K, Govindan R. Prognostic impact of malignant pleural effusion at presentation in patients with metastatic non-small-cell lung cancer. J Thorac Oncol. 2012; 7:1485–9.
Article91. William WN Jr, Lin HY, Lee JJ, Lippman SM, Roth JA, Kim ES. Revisiting stage IIIB and IV non-small cell lung cancer: analysis of the surveillance, epidemiology, and end results data. Chest. 2009; 136:701–9.92. Hooper C, Lee YC, Maskell N; Group BTSPG. Investigation of a unilateral pleural effusion in adults: British Thoracic Society Pleural Disease Guideline 2010. Thorax. 2010; 65 Suppl 2:ii4–17.
Article93. Lin J, Gu Y, Du R, Deng M, Lu Y, Ding Y. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non-small cell lung cancer patients by high resolution melting analysis and sequencing. Int J Clin Exp Pathol. 2014; 7:8813–22.94. Shin S, Kim J, Kim Y, Cho SM, Lee KA. Assessment of real-time PCR method for detection of EGFR mutation using both supernatant and cell pellet of malignant pleural effusion samples from non-small-cell lung cancer patients. Clin Chem Lab Med. 2017; 55:1962–9.
Article95. Qu X, Li Q, Yang J, et al. Double-stranded DNA in exosomes of malignant pleural effusions as a novel DNA source for EGFR mutation detection in lung adenocarcinoma. Front Oncol. 2019; 9:931.
Article96. Lee JS, Hur JY, Kim IA, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer. 2018; 18:1236.
Article97. Domagala-Kulawik J. The relevance of bronchoalveolar lavage fluid analysis for lung cancer patients. Expert Rev Respir Med. 2020; 14:329–37.
Article98. Miller RJ, Casal RF, Lazarus DR, Ost DE, Eapen GA. Flexible bronchoscopy. Clin Chest Med. 2018; 39:1–16.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Trends in Developing Extracellular Vesicle-Based Therapeutics
- Current Status and Prospects of Liquid Biopsy for Hepatocellular Carcinoma
- Establishment of External Quality Assessment Material Preparation Method for Next-Generation Sequencing-Based Liquid Biopsy Scheme
- Clinical Application of Circulating Tumor DNA Analysis
- Reported Lung Adenocarcinoma with Idiopathic Pulmonary Fibrosis after Open Lung Biopsy