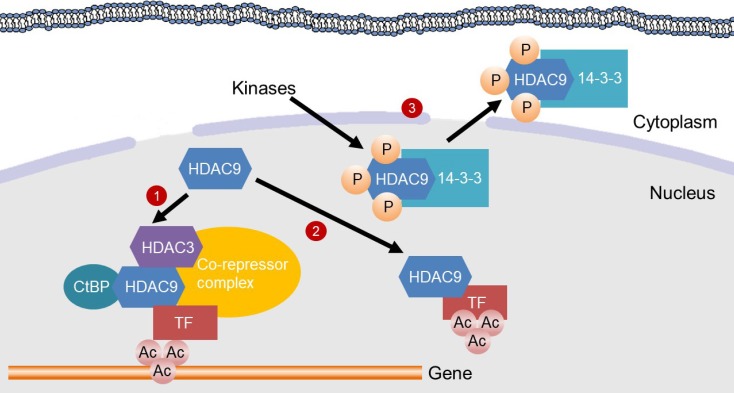
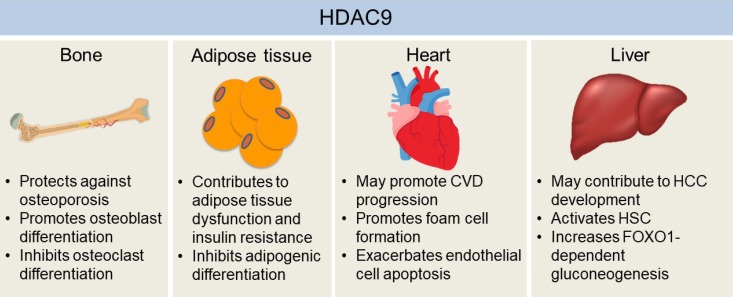
Diabetes Metab J.
2020 Apr;44(2):234-244. 10.4093/dmj.2019.0243.
Histone Deacetylase 9: Its Role in the Pathogenesis of Diabetes and Other Chronic Diseases
- Affiliations
-
- 1Department of Nutritional Sciences, University of Connecticut, Storrs, CT, USA
- 2Department of Internal Medicine, Kangwon National University School of Medicine, Chuncheon, Korea
- KMID: 2502404
- DOI: http://doi.org/10.4093/dmj.2019.0243
Abstract
- As a member of the class IIa histone deacetylases (HDACs), HDAC9 catalyzes the deacetylation of histones and transcription factors, commonly leading to the suppression of gene transcription. The activity of HDAC9 is regulated transcriptionally and posttranslationally. HDAC9 is known to play an essential role in regulating myocyte and adipocyte differentiation and cardiac muscle development. Also, recent studies have suggested that HDAC9 is involved in the pathogenesis of chronic diseases, including cardiovascular diseases, osteoporosis, autoimmune disease, cancer, obesity, insulin resistance, and liver fibrosis. HDAC9 modulates the expression of genes related to the pathogenesis of chronic diseases by altering chromatin structure in their promotor region or reducing the transcriptional activity of their respective transcription factors. This review summarizes the current knowledge of the regulation of HDAC9 expression and activity. Also, the roles of HDAC9 in the pathogenesis of chronic diseases are discussed, along with potential underlying mechanisms.
Keyword
Figure
Reference
-
1. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. PMID: 21321607.
Article2. Singh BN, Zhang G, Hwa YL, Li J, Dowdy SC, Jiang SW. Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther. 2010; 10:935–954. PMID: 20553216.
Article3. Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008; 9:206–218. PMID: 18292778.
Article4. de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003; 370:737–749. PMID: 12429021.5. Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci U S A. 2001; 98:10572–10577. PMID: 11535832.
Article6. Petrie K, Guidez F, Howell L, Healy L, Waxman S, Greaves M, Zelent A. The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem. 2003; 278:16059–16072. PMID: 12590135.
Article7. Zhou X, Richon VM, Rifkind RA, Marks PA. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc Natl Acad Sci U S A. 2000; 97:1056–1061. PMID: 10655483.
Article8. Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009; 10:32–42. PMID: 19065135.
Article9. Chatterjee TK, Idelman G, Blanco V, Blomkalns AL, Piegore MG Jr, Weintraub DS, Kumar S, Rajsheker S, Manka D, Rudich SM, Tang Y, Hui DY, Bassel-Duby R, Olson EN, Lingrel JB, Ho SM, Weintraub NL. Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J Biol Chem. 2011; 286:27836–27847. PMID: 21680747.
Article10. Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: conducting development and differentiation. Int J Dev Biol. 2009; 53:291–301. PMID: 19412888.
Article11. Han X, Han X, Wang Z, Shen J, Dong Q. HDAC9 regulates ox-LDL-induced endothelial cell apoptosis by participating in inflammatory reactions. Front Biosci (Landmark Ed). 2016; 21:907–917. PMID: 27100479.12. Jin Z, Wei W, Huynh H, Wan Y. HDAC9 inhibits osteoclastogenesis via mutual suppression of PPARγ/RANKL signaling. Mol Endocrinol. 2015; 29:730–738. PMID: 25793404.
Article13. Yan K, Cao Q, Reilly CM, Young NL, Garcia BA, Mishra N. Histone deacetylase 9 deficiency protects against effector T cell-mediated systemic autoimmunity. J Biol Chem. 2011; 286:28833–28843. PMID: 21708950.
Article14. Huang Y, Jian W, Zhao J, Wang G. Overexpression of HDAC9 is associated with poor prognosis and tumor progression of breast cancer in Chinese females. Onco Targets Ther. 2018; 11:2177–2184. PMID: 29713186.15. Greer CB, Tanaka Y, Kim YJ, Xie P, Zhang MQ, Park IH, Kim TH. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 2015; 13:1444–1455. PMID: 26549458.
Article16. Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. PMID: 19698979.
Article17. Chatterjee TK, Basford JE, Yiew KH, Stepp DW, Hui DY, Weintraub NL. Role of histone deacetylase 9 in regulating adipogenic differentiation and high fat diet-induced metabolic disease. Adipocyte. 2014; 3:333–338. PMID: 26317058.
Article18. Lehmann LH, Worst BC, Stanmore DA, Backs J. Histone deacetylase signaling in cardioprotection. Cell Mol Life Sci. 2014; 71:1673–1690. PMID: 24310814.
Article19. Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002; 9:45–57. PMID: 11804585.
Article20. Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001; 276:35826–35835. PMID: 11466315.21. Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007; 104:17335–17340. PMID: 17956988.
Article22. Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, Everett LJ, Nabel CS, Li Y, Selvakumaran V, Won KJ, Lazar MA. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013; 52:769–782. PMID: 24268577.
Article23. Parra M. Class IIa HDACs: new insights into their functions in physiology and pathology. FEBS J. 2015; 282:1736–1744. PMID: 25244360.24. McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001; 11:497–504. PMID: 11532390.
Article25. Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci. 2005; 8:313–321. PMID: 15711539.
Article26. Zhang CL, McKinsey TA, Lu JR, Olson EN. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2001; 276:35–39. PMID: 11022042.
Article27. Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009; 136:1056–1072. PMID: 19303849.
Article28. Griffin MJ, Wong RH, Pandya N, Sul HS. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J Biol Chem. 2007; 282:5453–5467. PMID: 17197698.
Article29. Mahmud SA, Manlove LS, Farrar MA. Interleukin-2 and STAT5 in regulatory T cell development and function. JAKSTAT. 2013; 2:e23154. PMID: 24058794.
Article30. Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal. 2012; 5:ra45. PMID: 22715468.
Article31. Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011; 146:1016–1028. PMID: 21925322.
Article32. Caron C, Boyault C, Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays. 2005; 27:408–415. PMID: 15770681.
Article33. Caron MM, Emans PJ, Cremers A, Surtel DA, Coolsen MM, van Rhijn LW, Welting TJ. Hypertrophic differentiation during chondrogenic differentiation of progenitor cells is stimulated by BMP-2 but suppressed by BMP-7. Osteoarthritis Cartilage. 2013; 21:604–613. PMID: 23353668.
Article34. Choi HJ, Kwon S, Kim DW. A post-translational modification cascade employing HDAC9-PIASy-RNF4 axis regulates chondrocyte hypertrophy by modulating Nkx3.2 protein stability. Cell Signal. 2016; 28:1336–1348. PMID: 27312341.
Article35. Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. EMBO J. 2003; 22:893–904. PMID: 12574125.
Article36. Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of beta-catenin by CREB-binding protein (CBP). J Biol Chem. 2002; 277:25562–25567. PMID: 11973335.37. Haberland M, Arnold MA, McAnally J, Phan D, Kim Y, Olson EN. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol Cell Biol. 2007; 27:518–525. PMID: 17101791.
Article38. Di Giorgio E, Brancolini C. Regulation of class IIa HDAC activities: it is not only matter of subcellular localization. Epigenomics. 2016; 8:251–269. PMID: 26791815.
Article39. McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001; 21:6312–6321. PMID: 11509672.
Article40. Miyamoto Y, Yamada K, Yoneda Y. Importin α: a key molecule in nuclear transport and non-transport functions. J Biochem. 2016; 160:69–75. PMID: 27289017.
Article41. Deng X, Ewton DZ, Mercer SE, Friedman E. Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J Biol Chem. 2005; 280:4894–4905. PMID: 15546868.
Article42. Harrison BC, Huynh K, Lundgaard GL, Helmke SM, Perryman MB, McKinsey TA. Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 2010; 584:1103–1110. PMID: 20188095.
Article43. Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007; 26:5450–5467. PMID: 17694086.
Article44. Muslin AJ, Xing H. 14-3-3 Proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000; 12:703–709. PMID: 11152955.
Article45. Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002; 110:479–488. PMID: 12202037.
Article46. International Stroke Genetics Consortium (ISGC). Wellcome Trust Case Control Consortium 2 (WTCCC2). Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, Jackson CA, Traylor M, Strange A, Su Z, Band G, Syme PD, Malik R, Pera J, Norrving B, Lemmens R, Freeman C, Schanz R, James T, Poole D, Murphy L, Segal H, Cortellini L, Cheng YC, Woo D, Nalls MA, Muller-Myhsok B, Meisinger C, Seedorf U, Ross-Adams H, Boonen S, Wloch-Kopec D, Valant V, Slark J, Furie K, Delavaran H, Langford C, Deloukas P, Edkins S, Hunt S, Gray E, Dronov S, Peltonen L, Gretarsdottir S, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boncoraglio GB, Parati EA, Attia J, Holliday E, Levi C, Franzosi MG, Goel A, Helgadottir A, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Duncanson A, Jankowski J, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Worrall BB, Kittner SJ, Mitchell BD, Kissela B, Meschia JF, Thijs V, Lindgren A, Macleod MJ, Slowik A, Walters M, Rosand J, Sharma P, Farrall M, Sudlow CL, Rothwell PM, Dichgans M, Donnelly P, Markus HS. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012; 44:328–333. PMID: 22306652.
Article47. Qingxu G, Yan Z, Jiannan X, Yunlong L. Association between the gene polymorphisms of HDAC9 and the risk of atherosclerosis and ischemic stroke. Pathol Oncol Res. 2016; 22:103–107. PMID: 26347468.
Article48. Azghandi S, Prell C, van der Laan SW, Schneider M, Malik R, Berer K, Gerdes N, Pasterkamp G, Weber C, Haffner C, Dichgans M. Deficiency of the stroke relevant HDAC9 gene attenuates atherosclerosis in accord with allele-specific effects at 7p21.1. Stroke. 2015; 46:197–202. PMID: 25388417.49. Cao Q, Rong S, Repa JJ, St Clair R, Parks JS, Mishra N. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arterioscler Thromb Vasc Biol. 2014; 34:1871–1879. PMID: 25035344.
Article50. Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007; 13:1496–1503. PMID: 18059282.51. Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, Liao EY, Luo XH. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015; 125:1509–1522. PMID: 25751060.
Article52. Chen YH, Yeh FL, Yeh SP, Ma HT, Hung SC, Hung MC, Li LY. Myocyte enhancer factor-2 interacting transcriptional repressor (MITR) is a switch that promotes osteogenesis and inhibits adipogenesis of mesenchymal stem cells by inactivating peroxisome proliferator-activated receptor gamma-2. J Biol Chem. 2011; 286:10671–10680. PMID: 21247904.53. Sun H, Kim JK, Mortensen R, Mutyaba LP, Hankenson KD, Krebsbach PH. Osteoblast-targeted suppression of PPARγ increases osteogenesis through activation of mTOR signaling. Stem Cells. 2013; 31:2183–2192. PMID: 23766271.
Article54. Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007; 13:1299–1307. PMID: 17922010.
Article55. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003; 4:330–336. PMID: 12612578.
Article56. de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010; 138:583–594. PMID: 19879272.
Article57. Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, Koike MA, Hancock SA, Bhatti TR, Han R, Jiao J, Veasey SC, Sims CA, Baur JA, Wallace DC, Hancock WW. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 2015; 29:2315–2326. PMID: 25681462.58. Salgado E, Bian X, Feng A, Shim H, Liang Z. HDAC9 overexpression confers invasive and angiogenic potential to triple negative breast cancer cells via modulating microRNA-206. Biochem Biophys Res Commun. 2018; 503:1087–1091. PMID: 29936177.
Article59. Liang Z, Bian X, Shim H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer. Biochem Biophys Res Commun. 2016; 477:461–466. PMID: 27318091.
Article60. Zheng Y, Yin L, Chen H, Yang S, Pan C, Lu S, Miao M, Jiao B. miR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012; 586:2396–2403. PMID: 22684007.
Article61. Zheng Y, Chen H, Yin M, Ye X, Chen G, Zhou X, Yin L, Zhang C, Ding B. MiR-376a and histone deacetylation 9 form a regulatory circuitry in hepatocellular carcinoma. Cell Physiol Biochem. 2015; 35:729–739. PMID: 25613642.
Article62. Chatterjee TK, Basford JE, Knoll E, Tong WS, Blanco V, Blomkalns AL, Rudich S, Lentsch AB, Hui DY, Weintraub NL. HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high-fat feeding. Diabetes. 2014; 63:176–187. PMID: 24101673.
Article63. Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008; 27:2320–2336. PMID: 18391974.
Article64. Zou Y, Gong N, Cui Y, Wang X, Cui A, Chen Q, Jiao T, Dong X, Yang H, Zhang S, Fang F, Chang Y. Forkhead box P1 (FOXP1) transcription factor regulates hepatic glucose homeostasis. J Biol Chem. 2015; 290:30607–30615. PMID: 26504089.
Article65. Chen J, Wang N, Dong M, Guo M, Zhao Y, Zhuo Z, Zhang C, Chi X, Pan Y, Jiang J, Tang H, Niu J, Yang D, Li Z, Han X, Wang Q, Chen X. The metabolic regulator histone deacetylase 9 contributes to glucose homeostasis abnormality induced by hepatitis C virus infection. Diabetes. 2015; 64:4088–4098. PMID: 26420860.
Article66. Chen J, Zhang Z, Wang N, Guo M, Chi X, Pan Y, Jiang J, Niu J, Ksimu S, Li JZ, Chen X, Wang Q. Role of HDAC9-FoxO1 axis in the transcriptional program associated with hepatic gluconeogenesis. Sci Rep. 2017; 7:6102. PMID: 28733598.
Article67. Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011; 145:607–621. PMID: 21565617.
Article68. Walkinshaw DR, Weist R, Kim GW, You L, Xiao L, Nie J, Li CS, Zhao S, Xu M, Yang XJ. The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J Biol Chem. 2013; 288:9345–9362. PMID: 23393134.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Pathogenesis and Pathophysiology of Chronic Obstructive Pulmonary Disease (COPD)
- The Role of Histone Acetylation in Mesenchymal Stem Cell Differentiation
- Roles of Histone Deacetylase 4 in the Inflammatory and Metabolic Processes
- HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases
- Activation of ATM-dependent DNA Damage Signal Pathway by a Histone Deacetylase Inhibitor, Trichostatin A