Cancer Res Treat.
2020 Jan;52(1):41-50. 10.4143/crt.2019.036.
Clinical Targeted Next-Generation sequencing Panels for Detection of Somatic Variants in Gliomas
- Affiliations
-
- 1Institute for Refractory Cancer Research, Samsung Medical Center, Seoul, Korea
- 2Deparment of Health Sciences and Technology, SAIHST, Sungkyunkwan University, Seoul, Korea
- 3Research Institute for Future Medicine, Samsung Medical Center, Seoul, Korea
- 4Samsung Genome Institute, Samsung Medical Center, Seoul, Korea
- 5Department of Neurosurgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 6Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2501200
- DOI: http://doi.org/10.4143/crt.2019.036
Abstract
- Purpose
Targeted next-generation sequencing (NGS) panels for solid tumors have been useful in clinical framework for accurate tumor diagnosis and identifying essential molecular aberrations. However, most cancer panels have been designed to address a wide spectrum of pan-cancer models, lacking integral prognostic markers that are highly specific to gliomas.
Materials and Methods
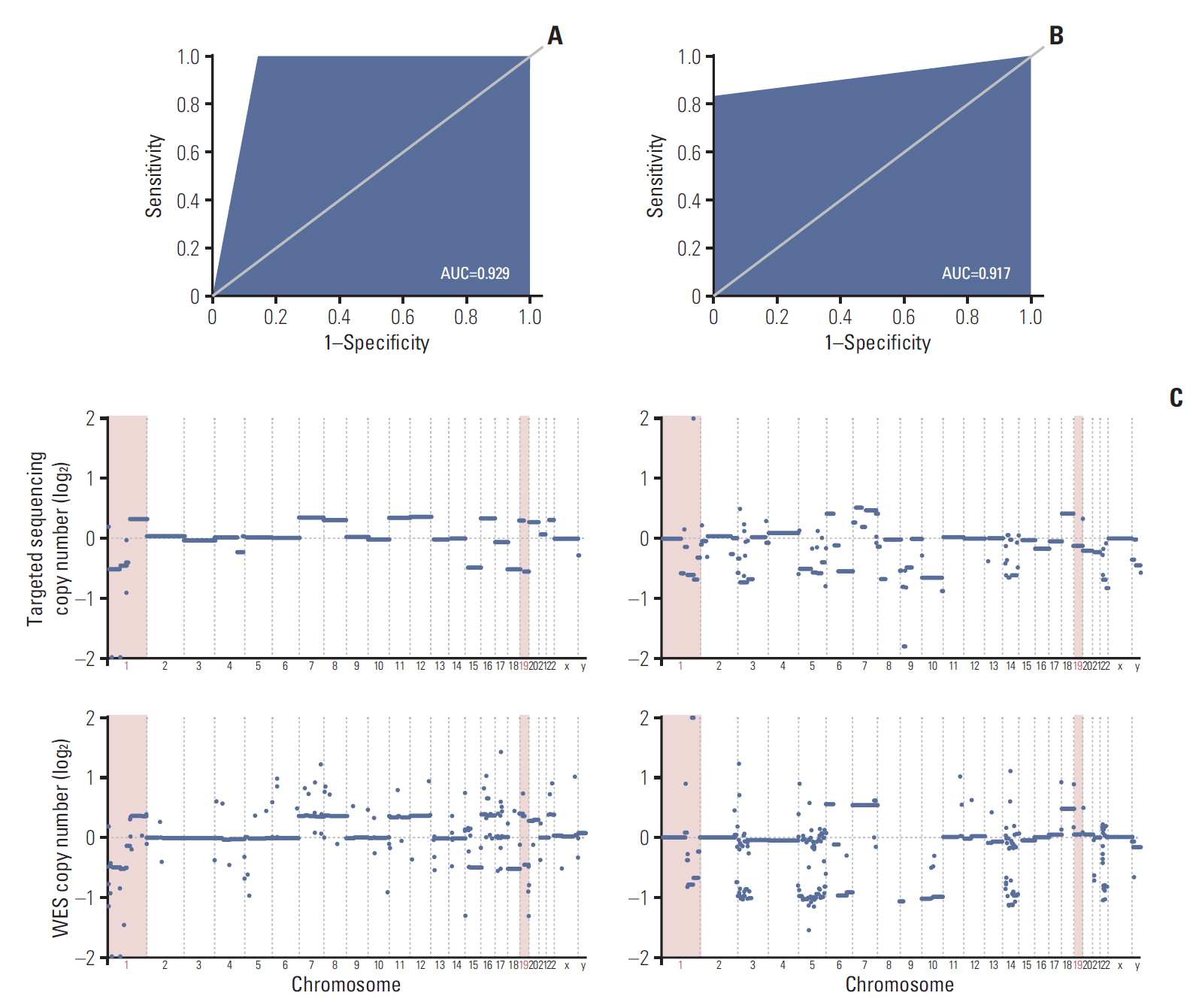
To address such challenges, we have developed a glioma-specific NGS panel, termed “GliomaSCAN,” that is capable of capturing single nucleotide variations and insertion/deletion, copy number variation, and selected promoter mutations and structural variations that cover a subset of intron regions in 232 essential glioma-associated genes. We confirmed clinical concordance rate using pairwise comparison of the identified variants from whole exome sequencing (WES), immunohistochemical analysis, and fluorescence in situ hybridization.
Results
Our panel demonstrated high sensitivity in detecting potential genomic variants that were present in the standard materials. To ensure the accuracy of our targeted sequencing panel, we compared our targeted panel to WES. The comparison results demonstrated a high correlation. Furthermore, we evaluated clinical utility of our panel in 46 glioma patients to assess the detection capacity of potential actionable mutations. Thirty-two patients harbored at least one recurrent somatic mutation in clinically actionable gene.
Conclusion
We have established a glioma-specific cancer panel. GliomaSCAN highly excelled in capturing somatic variations in terms of both sensitivity and specificity and provided potential clinical implication in facilitating genome-based clinical trials. Our results could provide conceptual advance towards improving the response of genomically guided molecularly targeted therapy in glioma patients.
Keyword
Figure
Reference
-
References
1. Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: a guerilla war. Acta Neuropathol. 2007; 114:443–58.
Article2. Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 2014; 128:551–9.
Article3. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131:803–20.
Article4. Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013; 98:E1852–60.
Article5. Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal evolution of the primary glioblastoma Genome. Cancer Cell. 2015; 28:318–28.
Article6. Liu Q, Nguyen DH, Dong Q, Shitaku P, Chung K, Liu OY, et al. Molecular properties of CD133+ glioblastoma stem cells derived from treatment-refractory recurrent brain tumors. J Neurooncol. 2009; 94:1–19.
Article7. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013; 31:213–9.
Article8. Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012; 486:405–9.
Article9. McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010; 26:2069–70.
Article10. Regnier MA, Raux M, Le Manach Y, Asencio Y, Gaillard J, Devilliers C, et al. Prognostic significance of blood lactate and lactate clearance in trauma patients. Anesthesiology. 2012; 117:1276–88.11. Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006; 27:861–74.
Article12. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–9.
Article13. Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014; 32:246–51.
Article14. Inda MM, Bonavia R, Seoane J. Glioblastoma multiforme: a look inside its heterogeneous nature. Cancers (Basel). 2014; 6:226–39.
Article15. Carr TH, McEwen R, Dougherty B, Johnson JH, Dry JR, Lai Z, et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer. 2016; 16:319–29.
Article16. Shen C, Meric-Bernstam F, Su X, Mendelsohn J, Giordano S. Prevalence of actionable mutations and copy number alterations and the price of a genomic testing panel. Oncotarget. 2016; 7:71686–95.
Article17. Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015; 121:631–9.
Article18. Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016; 48:768–76.
Article19. Wang YY, Zhang T, Li SW, Qian TY, Fan X, Peng XX, et al. Mapping p53 mutations in low-grade glioma: a voxel-based neuroimaging analysis. AJNR Am J Neuroradiol. 2015; 36:70–6.
Article20. Stander M, Peraud A, Leroch B, Kreth FW. Prognostic impact of TP53 mutation status for adult patients with supratentorial World Health Organization Grade II astrocytoma or oligoastrocytoma: a long-term analysis. Cancer. 2004; 101:1028–35.21. Rogers TW, Toor G, Drummond K, Love C, Field K, Asher R, et al. The 2016 revision of the WHO Classification of Central Nervous System Tumours: retrospective application to a cohort of diffuse gliomas. J Neurooncol. 2018; 137:181–9.
Article22. Levidou G, El-Habr E, Saetta AA, Bamias C, Katsouyanni K, Patsouris E, et al. P53 immunoexpression as a prognostic marker for human astrocytomas: a meta-analysis and review of the literature. J Neurooncol. 2010; 100:363–71.
Article23. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010; 17:98–110.
Article24. Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel). 2017; 9:E52.
Article25. Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006; 66:9852–61.
Article26. Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998; 90:1473–9.
Article27. Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for "Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV". Acta Neuropathol. 2018; 136:805–10.
Article28. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114:97–109.
Article29. Kim ST, Kim KM, Kim NK, Park JO, Ahn S, Yun JW, et al. Clinical application of targeted deep sequencing in solid-cancer patients and utility for biomarker-selected clinical trials. Oncologist. 2017; 22:1169–77.
Article30. Lee H, Lee KW, Lee T, Park D, Chung J, Lee C, et al. Performance evaluation method for read mapping tool in clinical panel sequencing. Genes Genomics. 2018; 40:189–97.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Korean Society for Genetic Diagnostics Guidelines for Validation of Next-Generation Sequencing-Based Somatic Variant Detection in Hematologic Malignancies
- Report of the Korean Association of External Quality Assessment Service on Next-Generation Sequencing Analysis for Somatic Variants (2018–2020)
- Targeted Next-Generation Sequencing of Plasma CellFree DNA in Korean Patients with Hepatocellular Carcinoma
- Detection of Mosaic Sequence Variants Associated with Human Genetic Diseases
- Pharmacogenomic Testing in the Clinical Laboratory: Historical Progress and Future Opportunities