J Breast Cancer.
2019 Dec;22(4):548-561. 10.4048/jbc.2019.22.e55.
Prognostic Role of KRAS mRNA Expression in Breast Cancer
- Affiliations
-
- 1Department of Surgery, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul, Korea. kiterius@snu.ac.kr
- 2Department of Radiation Oncology, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul, Korea.
- 3Department of Biostatistics, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul, Korea.
- 4Department of Pathology, Seoul National University College of Medicine, Seoul, Korea.
- 5Department of Surgery, Seoul Medical Center, Seoul, Korea.
- 6Department of Internal Medicine, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul, Korea.
- 7Department of Surgery, Graduate School, Kyung Hee University, Seoul, Korea.
- 8College of Pharmacy, Sookmyung Women's University, Seoul, Korea.
- KMID: 2470892
- DOI: http://doi.org/10.4048/jbc.2019.22.e55
Abstract
- PURPOSE
We investigated the prognostic role of KRAS mRNA expression in breast cancer using The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) databases.
METHODS
Clinical and biological data of 1,093 breast cancers from TCGA database and 1,904 breast cancers from METABRIC database were analyzed. Overall survival (OS) and breast cancer-specific survival (BCSS) were determined.
RESULTS
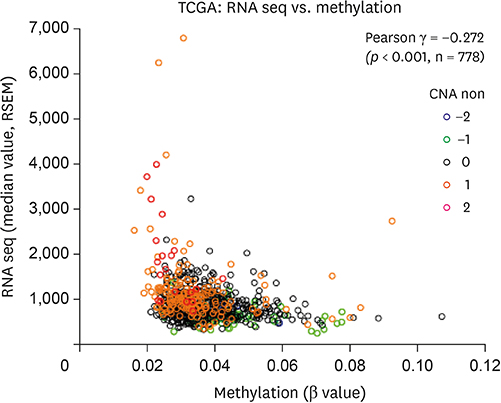
The group with high KRAS mRNA expression showed worse survival than the group with low KRAS mRNA expression regarding both OS (p = 0.012 in TCGA, p < 0.001 in METABRIC) and BCSS (p = 0.001 in METABRIC). According to multivariate analysis, the level of KRAS mRNA expression was an independent prognostic factor in both TCGA (hazard ratio [HR], 1.570; 95% confidence interval [CI], 1.026-2.403; p = 0.038) and METABRIC (HR, 1.254; 95% CI, 1.087-1.446; p = 0.002) databases. The prognostic impact of mRNA expression was effective only for luminal A subtype (p < 0.001 in METABRIC). Positive correlation was observed between mRNA expression and copy number alteration (CNA) (r = 0.577, p < 0.001 in TCGA; Ï = 0.343, p < 0.001 in METABRIC). Methylation showed negative correlations with both mRNA expression and CNA (r = −0.272, p < 0.001 in TCGA). The expression of mRNA had little association with the mutation status in breast cancers, having a mutation frequency of approximately 0.6%.
CONCLUSION
KRAS mRNA expression was significantly associated with breast cancer prognosis. It was found to be an independent prognostic factor for breast cancer. Prognostic role of KRAS mRNA expression was effective only in luminal A subtype. Further studies are needed to validate the prognostic role of KRAS mRNA expression in breast cancer, thus paving a way for clinical application of KRAS in practice.
Keyword
MeSH Terms
Figure
Reference
-
1. Goitre L, Trapani E, Trabalzini L, Retta SF. Chapter 1. The Ras superfamily of small GTPases: the unlocked secrets. In : Trabalzini L, Retta SF, editors. Ras Signaling: Methods and Protocols. Methods in Molecular Biology. Vol 1120. Totowa (NJ): Humana Press;2014. p. 1–18. DOI: 10.1007/978-1-62703-791-4.2. Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012; 491:399–405.3. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337.4. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–550.5. Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012; 72:2457–2467.
Article6. Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014; 13:828–851.
Article7. McGlynn LM, Kirkegaard T, Edwards J, Tovey S, Cameron D, Twelves C, et al. Ras/Raf-1/MAPK pathway mediates response to tamoxifen but not chemotherapy in breast cancer patients. Clin Cancer Res. 2009; 15:1487–1495.
Article8. Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The Genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018; 34:427–438.e6.
Article9. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.10. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015; 163:506–519.11. Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I, et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci U S A. 2012; 109:2730–2735.
Article12. Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010; 70:6509–6515.
Article13. Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011; 12:377–386.
Article14. Ovarian Cancer Association Consortium, Breast Cancer Association Consortium, and Consortium of Modifiers of BRCA1 and BRCA2. Hollestelle A, van der Baan FH, Berchuck A, Johnatty SE, Aben KK, et al. No clinical utility of KRAS variant rs61764370 for ovarian or breast cancer. Gynecol Oncol. 2016; 141:386–401.15. Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013; 45:1113–1120.
Article16. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012; 486:346–352.
Article17. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016; 7:11479.18. Harvey JJ. An unidentified virus which causes the rapid production of tumors in mice. Nature. 1964; 204:1104–1105.
Article19. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003; 3:459–465.
Article20. Kirsten WH, Mayer LA. Morphologic responses to a murine erythroblastosis virus. J Natl Cancer Inst. 1967; 39:311–335.21. Shih C, Weinberg RA. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell. 1982; 29:161–169.
Article22. Goldfarb M, Shimizu K, Perucho M, Wigler M. Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature. 1982; 296:404–409.
Article23. Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005; 118:843–846.
Article24. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013; 339:1546–1558.
Article25. Nagy Á, Pongor LS, Szabó A, Santarpia M, Győrffy B. KRAS driven expression signature has prognostic power superior to mutation status in non-small cell lung cancer. Int J Cancer. 2017; 140:930–937.
Article26. Wright KL, Adams JR, Liu JC, Loch AJ, Wong RG, Jo CE, et al. Ras signaling is a key determinant for metastatic dissemination and poor survival of luminal breast cancer patients. Cancer Res. 2015; 75:4960–4972.
Article27. Kim RK, Suh Y, Yoo KC, Cui YH, Kim H, Kim MJ, et al. Activation of KRAS promotes the mesenchymal features of basal-type breast cancer. Exp Mol Med. 2015; 47:e137.
Article28. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018; 173:321–337.e10.29. Sánchez-Muñoz A, Gallego E, de Luque V, Pérez-Rivas LG, Vicioso L, Ribelles N, et al. Lack of evidence for KRAS oncogenic mutations in triple-negative breast cancer. BMC Cancer. 2010; 10:136.
Article30. Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016; 129:1287–1292.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of High Mobility Group Protein Family [HMGI(Y) and HMGI-C] in Human Breast Cancer
- Vitamin D receptor (VDR) mRNA overexpression is associated with poor prognosis in breast carcinoma
- Relationship between Cathepsin D Expression and Other Prognostic Factors in Primary Breast Cancer
- Activation of KRAS promotes the mesenchymal features of basal-type breast cancer
- Expression of COX-2 in Cells of Invasive Ductal Carcinoma and Adjacent Non-Cancerous Ductal Epithelia in Human Breast