Ann Surg Treat Res.
2022 Oct;103(4):183-194. 10.4174/astr.2022.103.4.183.
Vitamin D receptor (VDR) mRNA overexpression is associated with poor prognosis in breast carcinoma
- Affiliations
-
- 1Department of Surgery, Konkuk University School of Medicine, Seoul, Korea
- 2Research Institute of Medical Science, Konkuk University School of Medicine, Seoul, Korea
- 3Department of Surgery, Konkuk University Medical Center, Seoul, Korea
- 4Department of Surgery, Kyung Hee University School of Medicine, Seoul, Korea
- KMID: 2534118
- DOI: http://doi.org/10.4174/astr.2022.103.4.183
Abstract
- Purpose
The prognostic value of vitamin D receptor gene (VDR) expression in breast cancer development is unclear. Here, we aimed to investigate whether VDR expression can be used as a prognostic indicator of breast cancer.
Methods
We used various public bioinformatics platforms: Oncomine, GEPIA, UALCAN, Kaplan-Meier plotter, UCSC XENA, bc-GenExMiner, WebGestalt, and STRING database.
Results
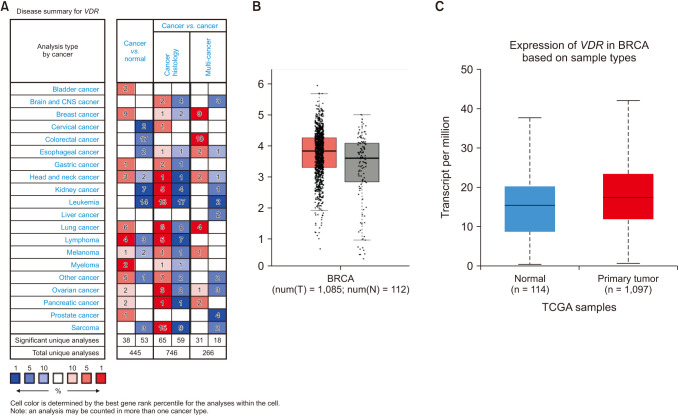
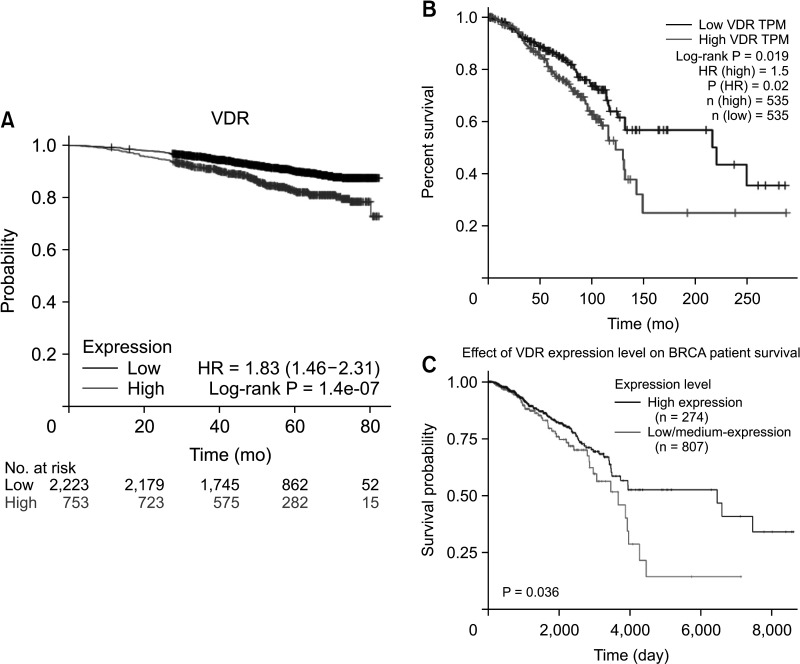
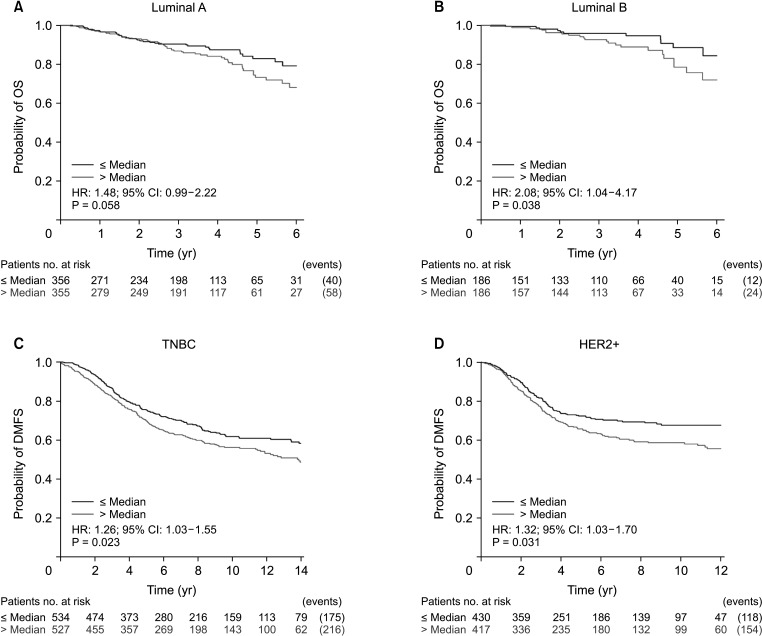
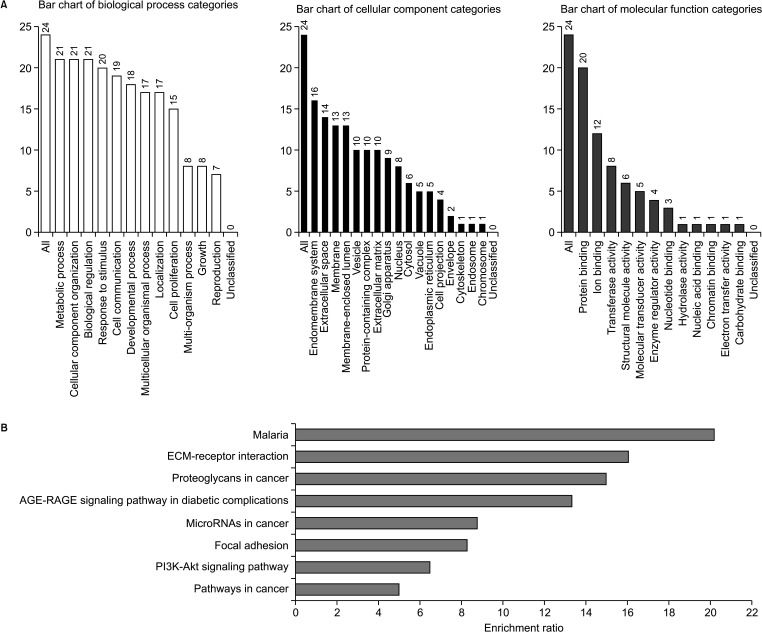
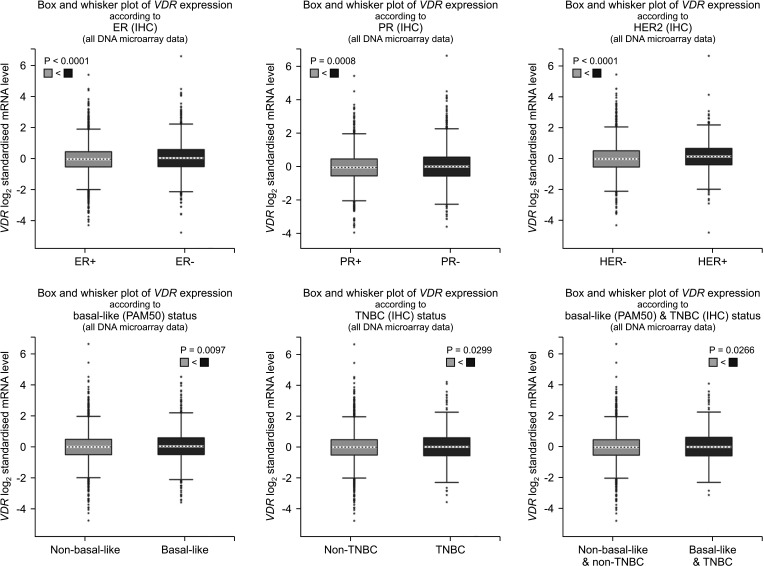
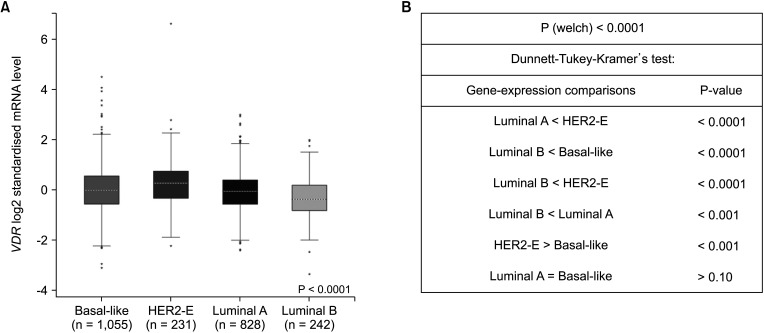
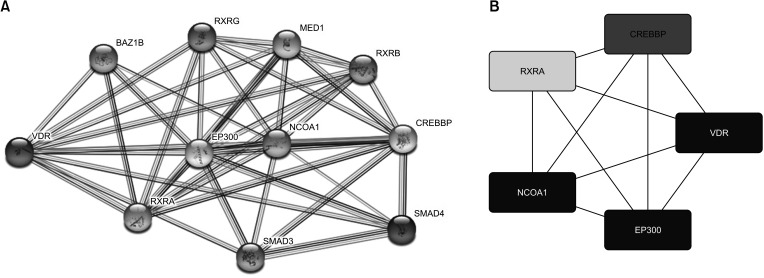
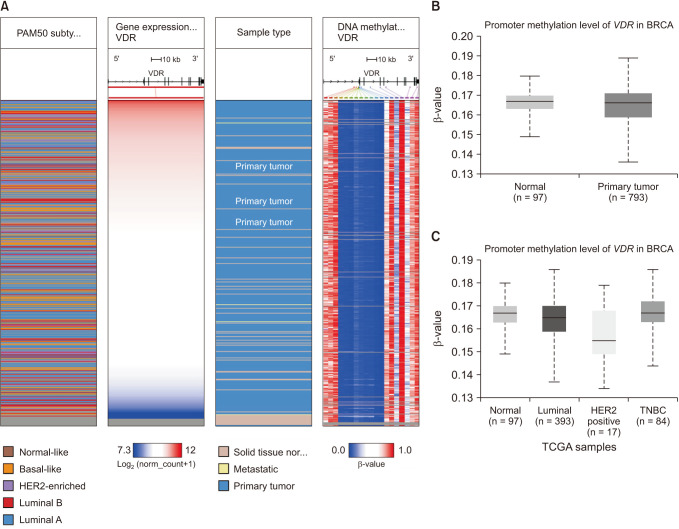
We found that VDR was upregulated in breast cancer in comparison to normal tissues. Overexpression of VDR was significantly associated with worse overall survival in breast cancer. The expression of VDR was related to age, TNM stages, estrogen receptor status, progesterone receptor status, human epidermal growth factor receptor 2 status, basallike (PAM 50) status, triple-negative breast cancer (TNBC) status, and basal-like (PAM 50) & TNBC status (P < 0.05). Increased VDR expression in breast cancer was significantly associated with older age. The 5 hub genes for VDR were NCOA1, EP300, CREBBP, and RXRA.
Conclusion
Our investigation offers hints about the prognostic role of VDR in breast cancer. The findings suggest that VDR expression might be used as a marker to determine a breast cancer patient’s prognosis. Nevertheless, further validation is warranted.
Figure
Reference
-
1. DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011; 61:409–418. PMID: 21969133.
Article2. Petracci E, Decarli A, Schairer C, Pfeiffer RM, Pee D, Masala G, et al. Risk factor modification and projections of absolute breast cancer risk. J Natl Cancer Inst. 2011; 103:1037–1048. PMID: 21705679.
Article3. Giovannucci E. The epidemiology of vitamin D and cancer incidence and mortality: a review (United States). Cancer Causes Control. 2005; 16:83–95. PMID: 15868450.
Article4. Keum N, Giovannucci E. Vitamin D supplements and cancer incidence and mortality: a meta-analysis. Br J Cancer. 2014; 111:976–980. PMID: 24918818.
Article5. Margolis RN, Christakos S. The nuclear receptor superfamily of steroid hormones and vitamin D gene regulation: an update. Ann N Y Acad Sci. 2010; 1192:208–214. PMID: 20392238.
Article6. Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007; 7:684–700. PMID: 17721433.
Article7. Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014; 14:342–357. PMID: 24705652.
Article8. Welsh J. Vitamin D and breast cancer: insights from animal models. Am J Clin Nutr. 2004; 80(6 Suppl):1721S–1724S. PMID: 15585794.
Article9. Anzano MA, Smith JM, Uskoković MR, Peer CW, Mullen LT, Letterio JJ, et al. 1 alpha,25-Dihydroxy-16-ene-23-yne-26,27-hexafluorocholecalciferol (Ro24-5531), a new deltanoid (vitamin D analogue) for prevention of breast cancer in the rat. Cancer Res. 1994; 54:1653–1656. PMID: 8137276.10. Crew KD, Gammon MD, Steck SE, Hershman DL, Cremers S, Dworakowski E, et al. Association between plasma 25-hydroxyvitamin D and breast cancer risk. Cancer Prev Res (Phila). 2009; 2:598–604. PMID: 19470790.
Article11. Murray A, Madden SF, Synnott NC, Klinger R, O’Connor D, O’Donovan N, et al. Vitamin D receptor as a target for breast cancer therapy. Endocr Relat Cancer. 2017; 24:181–195. PMID: 28213567.
Article12. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004; 6:1–6. PMID: 15068665.
Article13. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017; 45(W1):W98–W102. PMID: 28407145.
Article14. Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017; 19:649–658. PMID: 28732212.
Article15. Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014; 13:890–901. PMID: 24523301.
Article16. Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012; 131:765–775. PMID: 21452023.
Article17. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45(D1):D353–D361. PMID: 27899662.
Article18. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45(D1):D362–D368. PMID: 27924014.
Article19. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. PMID: 14597658.
Article20. Goldman M, Craft B, Zhu J, Haussler D. UCSC Xena for cancer genomics visualization and interpretation. Cancer Res. 2019; 79(13 Suppl):Abstract nr 911.21. Shaikh F, Baig S, Jamal Q. Do VDR gene polymorphisms contribute to breast cancer? Asian Pac J Cancer Prev. 2016; 17:479–483. PMID: 26925631.
Article22. Huss L, Butt ST, Borgquist S, Elebro K, Sandsveden M, Rosendahl A, et al. Vitamin D receptor expression in invasive breast tumors and breast cancer survival. Breast Cancer Res. 2019; 21:84. PMID: 31358030.
Article23. Al-Azhri J, Zhang Y, Bshara W, Zirpoli G, McCann SE, Khoury T, et al. Tumor expression of vitamin D receptor and breast cancer histopathological characteristics and prognosis. Clin Cancer Res. 2017; 23:97–103. PMID: 27407090.
Article24. Chakravarthi BV, Nepal S, Varambally S. Genomic and epigenomic alterations in cancer. Am J Pathol. 2016; 186:1724–1735. PMID: 27338107.
Article25. Herceg Z, Hainaut P. Genet ic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007; 1:26–41. PMID: 19383285.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Vitamin D receptor gene polymorphisms in breast cancer
- Upregulation of the Vitamin D Receptor in the Nasal Mucosa of Patients With Allergic Rhinitis
- c-erbB-2 Oncoprotein Overexpression in Breast Cancer
- Vitamin D Receptor Poly(A) Microsatellite Polymorphism and 25-Hydroxyvitamin D Serum Levels: Association with Susceptibility to Breast Cancer
- The Function of the Vitamin D Receptor and a Possible Role of Enhancer RNA in Epigenomic Regulation of Target Genes: Implications for Bone Metabolism