World J Mens Health.
2020 Jan;38(1):48-60. 10.5534/wjmh.190009.
Sex Differences in Gut Microbiota
- Affiliations
-
- 1Digestive Disease Research Institute and Department of Gastroenterology, Wonkwang University School of Medicine, Iksan, Korea. wms89@hanmail.net
- 2Division of Microbiome Research, VOWLIFE R&D Center, Iksan, Korea.
- 3Subtropical/Tropical Organism Gene Bank, Jeju National University, Jeju, Korea.
- 4Faculty of Biotechnology, School of Life Sciences, SARI, Jeju National University, Jeju, Korea.
- 5ChunLab, Inc., Seoul, Korea.
- 6Institute for Metabolic Disease, School of Medicine, Wonkwang University, Iksan, Korea.
- KMID: 2465409
- DOI: http://doi.org/10.5534/wjmh.190009
Abstract
- Humans carry numerous symbiotic microorganisms in their body, most of which are present in the gut. Although recent technological advances have produced extensive research data on gut microbiota, there are various confounding factors (e.g., diet, race, medications) to consider. Sex is one of the important variables affecting the gut microbiota, but the association has not yet been sufficiently investigated. Although the results are inconsistent, several animal and human studies have shown sex differences in gut microbiota. Herein, we review these studies to discuss the sex-dependent differences as well as the possible mechanisms involved.
MeSH Terms
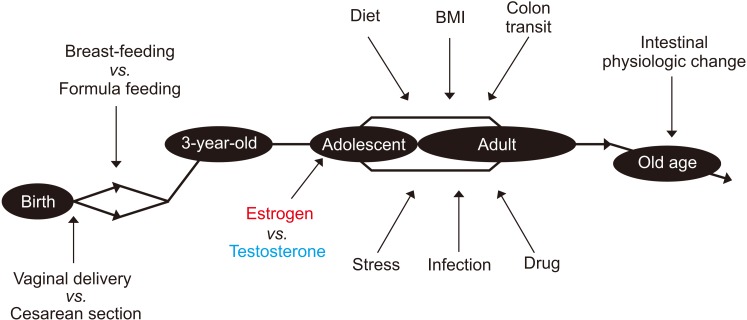
Figure
Cited by 1 articles
-
Sex- and Gender-related Issues of Gut Microbiota in Gastrointestinal Tract Diseases
Nayoung Kim
Korean J Gastroenterol. 2021;78(1):9-23. doi: 10.4166/kjg.2021.409.
Reference
-
1. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464:59–65. PMID: 20203603.
Article2. Sommer F, Bäckhed F. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol. 2013; 11:227–238. PMID: 23435359.3. Shanahan F. The colonic microbiota in health and disease. Curr Opin Gastroenterol. 2013; 29:49–54. PMID: 23041677.
Article4. Fukui H, Xu X, Miwa H. Role of gut microbiota-gut hormone axis in the pathophysiology of functional gastrointestinal disorders. J Neurogastroenterol Motil. 2018; 24:367–386. PMID: 29969855.
Article5. Osadchiy V, Martin CR, Mayer EA. The gut-brain axis and the microbiome: mechanisms and clinical implications. Clin Gastroenterol Hepatol. 2019; 17:322–332. PMID: 30292888.
Article6. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013; 39:400–412. PMID: 23973225.
Article7. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013; 339:1084–1088. PMID: 23328391.
Article8. Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016; 7:313–322. PMID: 27355107.
Article9. Lay C, Rigottier-Gois L, Holmstrøm K, Rajilic M, Vaughan EE, de Vos WM, et al. Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol. 2005; 71:4153–4155. PMID: 16000838.
Article10. Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006; 72:1027–1033. PMID: 16461645.
Article11. Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008; 105:2117–2122. PMID: 18252821.
Article12. Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014; 509:357–360. PMID: 24739969.
Article13. Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015; 10:e0124599. PMID: 25874569.
Article14. Singh P, Manning SD. Impact of age and sex on the composition and abundance of the intestinal microbiota in individuals with and without enteric infections. Ann Epidemiol. 2016; 26:380–385. PMID: 27118420.
Article15. Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016; 11:e0154090. PMID: 27228093.
Article16. Borgo F, Garbossa S, Riva A, Severgnini M, Luigiano C, Benetti A, et al. Body mass index and sex affect diverse microbial niches within the gut. Front Microbiol. 2018; 9:213. PMID: 29491857.
Article17. Gao X, Zhang M, Xue J, Huang J, Zhuang R, Zhou X, et al. Body mass index differences in the gut microbiota are gender specific. Front Microbiol. 2018; 9:1250. PMID: 29988340.
Article18. Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J Gastroenterol. 2019; 54:53–63. PMID: 29926167.
Article19. Sinha T, Vich Vila A, Garmaeva S, Jankipersadsing SA, Imhann F, Collij V, et al. Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes. 2019; 10:358–366. PMID: 30373468.
Article20. Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol. 2011; 61:423–428. PMID: 21181142.
Article21. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. PMID: 22699609.22. Kim YS. Sex and gender-related issues in biomedical science. Sci Ed. 2018; 5:66–69.
Article23. Jahng J, Kim YS. Why should we contemplate on gender difference in functional gastrointestinal disorders? J Neurogastroenterol Motil. 2017; 23:1–2. PMID: 28049860.
Article24. Lee H, Pak YK, Yeo EJ, Kim YS, Paik HY, Lee SK. It is time to integrate sex as a variable in preclinical and clinical studies. Exp Mol Med. 2018; 50:82. PMID: 30038313.
Article25. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007; 449:804–810. PMID: 17943116.
Article26. Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012; 18:1185–1193. PMID: 23033984.
Article27. Bilen M, Dufour JC, Lagier JC, Cadoret F, Daoud Z, Dubourg G, et al. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome. 2018; 6:94. PMID: 29793532.
Article28. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans? Cell. 2016; 164:337–340. PMID: 26824647.
Article29. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533. PMID: 27541692.
Article30. The Lancet Infectious Diseases. Microbiome studies and “blue whales in the Himalayas”. Lancet Infect Dis. 2018; 18:925. PMID: 30152348.31. Bischoff SC. Microbiota and aging. Curr Opin Clin Nutr Metab Care. 2016; 19:26–30. PMID: 26560527.32. Salles N. Basic mechanisms of the aging gastrointestinal tract. Dig Dis. 2007; 25:112–117. PMID: 17468545.
Article33. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180. PMID: 21508958.
Article34. Nishijima S, Suda W, Oshima K, Kim SW, Hirose Y, Morita H, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016; 23:125–133. PMID: 26951067.
Article35. Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, et al. Rethinking “enterotypes”. Cell Host Microbe. 2014; 16:433–437. PMID: 25299329.
Article36. Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, et al. US immigration westernizes the human gut microbiome. Cell. 2018; 175:962–972.e10. PMID: 30388453.
Article37. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017; 15:630–638. PMID: 28626231.
Article38. Fushuku S, Fukuda K. Gender difference in the composition of fecal flora in laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp Anim. 2008; 57:489–493. PMID: 18946187.
Article39. Elderman M, Hugenholtz F, Belzer C, Boekschoten M, van Beek A, de Haan B, et al. Sex and strain dependent differences in mucosal immunology and microbiota composition in mice. Biol Sex Differ. 2018; 9:26. PMID: 29914546.
Article40. Fransen F, van Beek AA, Borghuis T, Meijer B, Hugenholtz F, van der Gaast-de Jongh C, et al. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol. 2017; 8:754. PMID: 28713378.
Article41. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016; 352:560–564. PMID: 27126039.
Article42. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016; 352:565–569. PMID: 27126040.
Article43. Bernbom N, Nørrung B, Saadbye P, Mølbak L, Vogensen FK, Licht TR. Comparison of methods and animal models commonly used for investigation of fecal microbiota: effects of time, host and gender. J Microbiol Methods. 2006; 66:87–95. PMID: 16289391.
Article44. Chen KL, Madak-Erdogan Z. Estrogen and microbiota cross-talk: Should we pay attention? Trends Endocrinol Metab. 2016; 27:752–755. PMID: 27553057.
Article45. Cox-York KA, Sheflin AM, Foster MT, Gentile CL, Kahl A, Koch LG, et al. Ovariectomy results in differential shifts in gut microbiota in low versus high aerobic capacity rats. Physiol Rep. 2015; 3:e12488. PMID: 26265751.
Article46. Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012; 10:253. PMID: 23259758.
Article47. Nakatsu CH, Armstrong A, Clavijo AP, Martin BR, Barnes S, Weaver CM. Fecal bacterial community changes associated with isoflavone metabolites in postmenopausal women after soy bar consumption. PLoS One. 2014; 9:e108924. PMID: 25271941.
Article48. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018; 555:623–628. PMID: 29555994.
Article49. Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014; 5:4500. PMID: 25072318.
Article50. Shastri P, McCarville J, Kalmokoff M, Brooks SP, Green-Johnson JM. Sex differences in gut fermentation and immune parameters in rats fed an oligofructose-supplemented diet. Biol Sex Differ. 2015; 6:13. PMID: 26251695.
Article51. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011; 334:105–108. PMID: 21885731.
Article52. Gaskins AJ, Mumford SL, Zhang C, Wactawski-Wende J, Hovey KM, Whitcomb BW, et al. Effect of daily fiber intake on reproductive function: the BioCycle study. Am J Clin Nutr. 2009; 90:1061–1069. PMID: 19692496.53. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005; 102:11070–11075. PMID: 16033867.
Article54. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–1023. PMID: 17183309.55. Mestdagh R, Dumas ME, Rezzi S, Kochhar S, Holmes E, Claus SP, et al. Gut microbiota modulate the metabolism of brown adipose tissue in mice. J Proteome Res. 2012; 11:620–630. PMID: 22053906.
Article56. Szymczak J, Milewicz A, Thijssen JH, Blankenstein MA, Daroszewski J. Concentration of sex steroids in adipose tissue after menopause. Steroids. 1998; 63:319–321. PMID: 9618794.
Article57. Tottey W, Feria-Gervasio D, Gaci N, Laillet B, Pujos E, Martin JF, et al. Colonic transit time is a driven force of the gut microbiota composition and metabolism: in vitro evidence. J Neurogastroenterol Motil. 2017; 23:124–134. PMID: 27530163.
Article58. Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016; 65:57–62. PMID: 26069274.
Article59. Parthasarathy G, Chen J, Chen X, Chia N, O'Connor HM, Wolf PG, et al. Relationship between microbiota of the colonic mucosa vs feces and symptoms, colonic transit, and methane production in female patients with chronic constipation. Gastroenterology. 2016; 150:367–379.e1. PMID: 26460205.
Article60. Lampe JW, Fredstrom SB, Slavin JL, Potter JD. Sex differences in colonic function: a randomised trial. Gut. 1993; 34:531–536. PMID: 8387940.
Article61. Degen LP, Phillips SF. Variability of gastrointestinal transit in healthy women and men. Gut. 1996; 39:299–305. PMID: 8977347.
Article62. Meier R, Beglinger C, Dederding JP, Meyer-Wyss B, Fumagalli M, Rowedder A, et al. Influence of age, gender, hormonal status and smoking habits on colonic transit time. Neurogastroenterol Motil. 1995; 7:235–238. PMID: 8574912.
Article63. Song BK, Cho KO, Jo Y, Oh JW, Kim YS. Colon transit time according to physical activity level in adults. J Neurogastroenterol Motil. 2012; 18:64–69. PMID: 22323989.
Article64. Jung HK, Kim DY, Moon IH. Effects of gender and menstrual cycle on colonic transit time in healthy subjects. Korean J Intern Med. 2003; 18:181–186. PMID: 14619388.
Article65. Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil. 2014; 20:31–40. PMID: 24466443.
Article66. Danska JS. Sex matters for mechanism. Sci Transl Med. 2014; 6:258fs40.
Article67. Kim YS, Kim N. Sex-gender differences in irritable bowel syndrome. J Neurogastroenterol Motil. 2018; 24:544–558. PMID: 30347934.
Article68. Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009; 136:1979–1988. PMID: 19457422.
Article69. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016; 16:626–638. PMID: 27546235.
Article70. Elderman M, de Vos P, Faas M. Role of microbiota in sexually dimorphic immunity. Front Immunol. 2018; 9:1018. PMID: 29910797.
Article71. Wallis A, Butt H, Ball M, Lewis DP, Bruck D. Support for the microgenderome: associations in a human clinical population. Sci Rep. 2016; 6:19171. PMID: 26757840.
Article72. Kozik AJ, Nakatsu CH, Chun H, Jones-Hall YL. Age, sex, and TNF associated differences in the gut microbiota of mice and their impact on acute TNBS colitis. Exp Mol Pathol. 2017; 103:311–319. PMID: 29175304.
Article73. Lee JY, Kim N, Nam RH, Sohn SH, Lee SM, Choi D, et al. Probiotics reduce repeated water avoidance stress-induced colonic microinflammation in Wistar rats in a sex-specific manner. PLoS One. 2017; 12:e0188992. PMID: 29244820.
Article74. Karunasena E, McMahon KW, Chang D, Brashears MM. Host responses to the pathogen Mycobacterium avium subsp. paratuberculosis and beneficial microbes exhibit host sex specificity. Appl Environ Microbiol. 2014; 80:4481–4490. PMID: 24814797.
Article75. uBiome. A surprising comparison of male vs. female microbiomes [Internet]. San Francisco (CA): uBiome;c2015. cited 2018 Dec 10. Available from: https://ubiome.com/blog/post/surprising-comparison-male-vs-female-microbiomes/.