Korean J Physiol Pharmacol.
2019 Nov;23(6):529-537. 10.4196/kjpp.2019.23.6.529.
Expression of potassium channel genes predicts clinical outcome in lung cancer
- Affiliations
-
- 1Department of Physiology and Cell Biology, University of Nevada, Reno School of Medicine, Reno, Nevada 89557, USA.
- 2Department of Physiology, College of Medicine, Chung-Ang University, Seoul 06974, Korea. akdongyi01@cau.ac.kr
- 3Department of Family Medicine, Chung-Ang University Hospital, College of Medicine, Chung-Ang University, Seoul 06973, Korea.
- KMID: 2461046
- DOI: http://doi.org/10.4196/kjpp.2019.23.6.529
Abstract
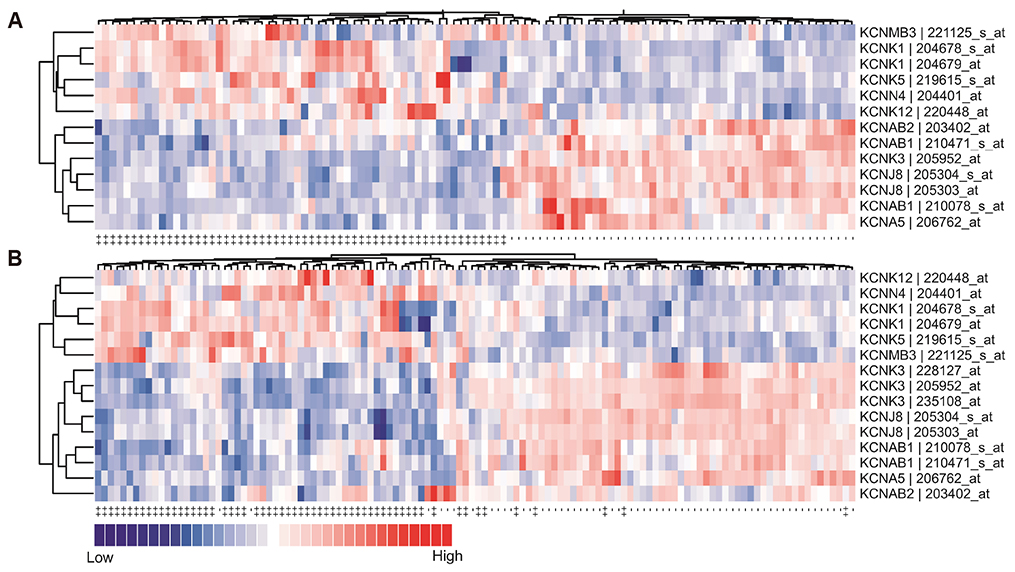
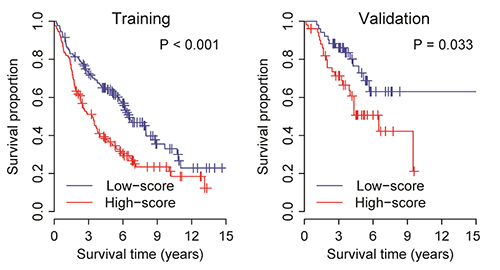
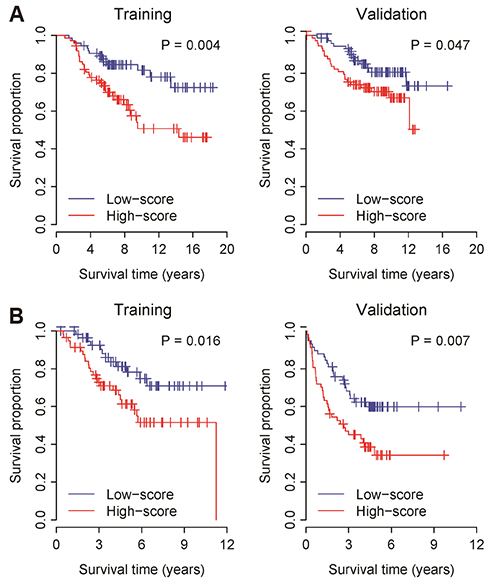
- Lung cancer is the most common cause of cancer deaths worldwide and several molecular signatures have been developed to predict survival in lung cancer. Increasing evidence suggests that proliferation and migration to promote tumor growth are associated with dysregulated ion channel expression. In this study, by analyzing high-throughput gene expression data, we identify the differentially expressed K⺠channel genes in lung cancer. In total, we prioritize ten dysregulated K⺠channel genes (5 up-regulated and 5 down-regulated genes, which were designated as K-10) in lung tumor tissue compared with normal tissue. A risk scoring system combined with the K-10 signature accurately predicts clinical outcome in lung cancer, which is independent of standard clinical and pathological prognostic factors including patient age, lymph node involvement, tumor size, and tumor grade. We further indicate that the K-10 potentially predicts clinical outcome in breast and colon cancers. Molecular signature discovered through K⺠gene expression profiling may serve as a novel biomarker to assess the risk in lung cancer.
MeSH Terms
Figure
Reference
-
1. Thorneloe KS, Nelson MT. Ion channels in smooth muscle: regulators of intracellular calcium and contractility. Can J Physiol Pharmacol. 2005; 83:215–242.
Article2. Krasznai Z. Ion channels in T cells: from molecular pharmacology to therapy. Arch Immunol Ther Exp (Warsz). 2005; 53:127–135.3. Mason WT, Rawlings SR, Cobbett P, Sikdar SK, Zorec R, Akerman SN, Benham CD, Berridge MJ, Cheek T, Moreton RB. Control of secretion in anterior pituitary cells--linking ion channels, messengers and exocytosis. J Exp Biol. 1988; 139:287–316.
Article4. Baker EH. Ion channels and the control of blood pressure. Br J Clin Pharmacol. 2000; 49:185–198.
Article5. Jentsch TJ, Hübner CA, Fuhrmann JC. Ion channels: function unravelled by dysfunction. Nat Cell Biol. 2004; 6:1039–1047.
Article6. Lehmann-Horn F, Jurkat-Rott K. Voltage-gated ion channels and hereditary disease. Physiol Rev. 1999; 79:1317–1372.
Article7. Hübner CA, Jentsch TJ. Ion channel diseases. Hum Mol Genet. 2002; 11:2435–2445.8. Curran ME. Potassium ion channels and human disease: phenotypes to drug targets? Curr Opin Biotechnol. 1998; 9:565–572.
Article9. Cooper EC, Jan LY. Ion channel genes and human neurological disease: recent progress, prospects, and challenges. Proc Natl Acad Sci U S A. 1999; 96:4759–4766.
Article10. Hanna MG, Wood NW, Kullmann DM. Ion channels and neurological disease: DNA based diagnosis is now possible, and ion channels may be important in common paroxysmal disorders. J Neurol Neurosurg Psychiatry. 1998; 65:427–431.
Article11. Bloch M, Ousingsawat J, Simon R, Schraml P, Gasser TC, Mihatsch MJ, Kunzelmann K, Bubendorf L. KCNMA1 gene amplification promotes tumor cell proliferation in human prostate cancer. Oncogene. 2007; 26:2525–2534.
Article12. Le Guennec JY, Ouadid-Ahidouch H, Soriani O, Besson P, Ahidouch A, Vandier C. Voltage-gated ion channels, new targets in anti-cancer research. Recent Pat Anticancer Drug Discov. 2007; 2:189–202.13. Fiske JL, Fomin VP, Brown ML, Duncan RL, Sikes RA. Voltage-sensitive ion channels and cancer. Cancer Metastasis Rev. 2006; 25:493–500.
Article14. Roger S, Potier M, Vandier C, Besson P, Le Guennec JY. Voltage-gated sodium channels: new targets in cancer therapy? Curr Pharm Des. 2006; 12:3681–3695.
Article15. Pardo LA, Stühmer W. Eag1: an emerging oncological target. Cancer Res. 2008; 68:1611–1613.16. Pardo LA, Contreras-Jurado C, Zientkowska M, Alves F, Stühmer M. Role of voltage-gated potassium channels in cancer. J Membr Biol. 2005; 205:115–124.
Article17. Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005; 205:159–173.
Article18. Gómez-Varela D, Zwick-Wallasch E, Knötgen H, Sánchez A, Hettmann T, Ossipov D, Weseloh R, Contreras-Jurado C, Rothe M, Stühmer W, Pardo LA. Monoclonal antibody blockade of the human Eag1 potassium channel function exerts antitumor activity. Cancer Res. 2007; 67:7343–7349.
Article19. Pillozzi S, Masselli M, De Lorenzo E, Accordi B, Cilia E, Crociani O, Amedei A, Veltroni M, D'Amico M, Basso G, Becchetti A, Campana D, Arcangeli A. Chemotherapy resistance in acute lymphoblastic leukemia requires hERG1 channels and is overcome by hERG1 blockers. Blood. 2011; 117:902–914.
Article20. Villalonga N, Ferreres JC, Argilés JM, Condom E, Felipe A. Potassium channels are a new target field in anticancer drug design. Recent Pat Anticancer Drug Discov. 2007; 2:212–223.
Article21. Li M, Xiong ZG. Ion channels as targets for cancer therapy. Int J Physiol Pathophysiol Pharmacol. 2011; 3:156–166.22. Suzuki T, Takimoto K. Selective expression of HERG and Kv2 channels influences proliferation of uterine cancer cells. Int J Oncol. 2004; 25:153–159.23. Brevet M, Haren N, Sevestre H, Merviel P, Ouadid-Ahidouch H. DNA methylation of Kv1.3 potassium channel gene promoter is associated with poorly differentiated breast adenocarcinoma. Cell Physiol Biochem. 2009; 24:25–32.
Article24. Basrai D, Kraft R, Bollensdorff C, Liebmann L, Benndorf K, Patt S. BK channel blockers inhibit potassium-induced proliferation of human astrocytoma cells. Neuroreport. 2002; 13:403–407.
Article25. Weaver AK, Liu X, Sontheimer H. Role for calcium-activated potassium channels (BK) in growth control of human malignant glioma cells. J Neurosci Res. 2004; 78:224–234.
Article26. Abdul M, Hoosein N. Expression and activity of potassium ion channels in human prostate cancer. Cancer Lett. 2002; 186:99–105.
Article27. Gessner G, Schönherr K, Soom M, Hansel A, Asim M, Baniahmad A, Derst C, Hoshi T, Heinemann SH. BKCa channels activating at resting potential without calcium in LNCaP prostate cancer cells. J Membr Biol. 2005; 208:229–240.
Article28. Voloshyna I, Besana A, Castillo M, Matos T, Weinstein IB, Mansukhani M, Robinson RB, Cordon-Cardo C, Feinmark SJ. TREK-1 is a novel molecular target in prostate cancer. Cancer Res. 2008; 68:1197–1203.
Article29. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127:2893–2917.
Article30. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61:69–90.
Article31. Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, Strumpf D, Hanash S, Shepherd FA, Ding K, Seymour L, Naoki K, Pennell N, Weir B, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008; 14:822–827.
Article32. Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008; 26:877–883.
Article33. Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, Tucker MA, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM, Wang E. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010; 16:430–441.
Article34. Pitroda SP, Zhou T, Sweis RF, Filippo M, Labay E, Beckett MA, Mauceri HJ, Liang H, Darga TE, Perakis S, Khan SA, Sutton HG, Zhang W, Khodarev NN, Garcia JG, Weichselbaum RR. Tumor endothelial inflammation predicts clinical outcome in diverse human cancers. PLoS One. 2012; 7:e46104.
Article35. Perumal D, Singh S, Yoder SJ, Bloom GC, Chellappan SP. A novel five gene signature derived from stem-like side population cells predicts overall and recurrence-free survival in NSCLC. PLoS One. 2012; 7:e43589.
Article36. Kossenkov AV, Dawany N, Evans TL, Kucharczuk JC, Albelda SM, Showe LC, Showe MK, Vachani A. Peripheral immune cell gene expression predicts survival of patients with non-small cell lung cancer. PLoS One. 2012; 7:e34392.
Article37. Sharman JL, Mpamhanga CP, Spedding M, Germain P, Staels B, Dacquet C, Laudet V, Harmar AJ. NC-IUPHAR. IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res. 2011; 39(Database issue):D534–D538.
Article38. van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002; 347:1999–2009.
Article39. Choi JK, Yu U, Kim S, Yoo OJ. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003; 19 Suppl 1:i84–i90.
Article40. Parmigiani G, Garrett ES, Anbazhagan R, Gabrielson E. A statistical framework for expression-based molecular classification in cancer. J R Stat Soc Series B Stat Methodol. 2002; 64:717–736.
Article41. R_Development_Core_Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing;2005.42. Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder S, Shih JH, Caporaso NE, Jen J. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008; 3:e1651.
Article43. Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, Chuang EY. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010; 19:2590–2597.
Article44. Breuer EK, Fukushiro-Lopes D, Dalheim A, Burnette M, Zartman J, Kaja S, Wells C, Campo L, Curtis KJ, Romero-Moreno R, Littlepage LE, Niebur GL, Hoskins K, Nishimura MI, Gentile S. Potassium channel activity controls breast cancer metastasis by affecting β-catenin signaling. Cell Death Dis. 2019; 10:180.
Article45. Ru Q, Tian X, Wu YX, Wu RH, Pi MS, Li CY. Voltage-gated and ATP-sensitive K+ channels are associated with cell proliferation and tumorigenesis of human glioma. Oncol Rep. 2014; 31:842–848.46. Sundelacruz S, Levin M, Kaplan DL. Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev Rep. 2009; 5:231–246.
Article47. Spitzner M, Ousingsawat J, Scheidt K, Kunzelmann K, Schreiber R. Voltage-gated K+ channels support proliferation of colonic carcinoma cells. FASEB J. 2007; 21:35–44.48. Jiang S, Zhu L, Yang J, Hu L, Gu J, Xing X, Sun Y, Zhang Z. Integrated expression profiling of potassium channels identifys KCNN4 as a prognostic biomarker of pancreatic cancer. Biochem Biophys Res Commun. 2017; 494:113–119.
Article49. Williams S, Bateman A, O'Kelly I. Altered expression of two-pore domain potassium (K2P) channels in cancer. PLoS One. 2013; 8:e74589.
Article50. Comes N, Bielanska J, Vallejo-Gracia A, Serrano-Albarrás A, Marruecos L, Gómez D, Soler C, Condom E, Ramón Y, Hernández-Losa J, Ferreres JC, Felipe A. The voltage-dependent K+ channels Kv1.3 and Kv1.5 in human cancer. Front Physiol. 2013; 4:283.51. Hauptman N, Jevšinek Skok D, Spasovska E, Boštjančič E, Glavač D. Genes CEP55, FOXD3, FOXF2, GNAO1, GRIA4, and KCNA5 as potential diagnostic biomarkers in colorectal cancer. BMC Med Genomics. 2019; 12:54.
Article52. Cheng YY, Wright CM, Kirschner MB, Williams M, Sarun KH, Sytnyk V, Leshchynska I, Edelman JJ, Vallely MP, McCaughan BC, Klebe S, van Zandwijk N, Lin RC, Reid G. KCa1.1, a calcium-activated potassium channel subunit alpha 1, is targeted by miR-17-5p and modulates cell migration in malignant pleural mesothelioma. Mol Cancer. 2016; 15:44.
Article53. Mu D, Chen L, Zhang X, See LH, Koch CM, Yen C, Tong JJ, Spiegel L, Nguyen KC, Servoss A, Peng Y, Pei L, Marks JR, Lowe S, Hoey T, Jan LY, McCombie WR, Wigler MH, Powers S. Genomic amplification and oncogenic properties of the KCNK9 potassium channel gene. Cancer Cell. 2003; 3:297–302.
Article54. Iwasa H, Kurabayashi M, Nagai R, Nakamura Y, Tanaka T. Genetic variations in five genes involved in the excitement of cardiomyocytes. J Hum Genet. 2001; 46:549–552.
Article55. Toncheva D, Mihailova-Hristova M, Vazharova R, Staneva R, Karachanak S, Dimitrov P, Simeonov V, Ivanov S, Balabanski L, Serbezov D, Malinov M, Stefanovic V, Čukuranovič R, Polenakovic M, Jankovic-Velickovic L, Djordjevic V, Jevtovic-Stoimenov T, Plaseska-Karanfilska D, Galabov A, Djonov V, et al. NGS nominated CELA1, HSPG2, and KCNK5 as candidate genes for predisposition to Balkan endemic nephropathy. Biomed Res Int. 2014; 2014:920723.56. Lorenz S, Heils A, Kasper JM, Sander T. Allelic association of a truncation mutation of the KCNMB3 gene with idiopathic generalized epilepsy. Am J Med Genet B Neuropsychiatr Genet. 2007; 144B:10–13.57. Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A, Terzic A. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006; 15:2185–2191.58. Remillard CV, Tigno DD, Platoshyn O, Burg ED, Brevnova EE, Conger D, Nicholson A, Rana BK, Channick RN, Rubin LJ, O'connor DT, Yuan JX. Function of Kv1.5 channels and genetic variations of KCNA5 in patients with idiopathic pulmonary arterial hypertension. Am J Physiol Cell Physiol. 2007; 292:C1837–C1853.59. Navas Tejedor P, Tenorio Castaño J, Palomino Doza J, Arias Lajara P, Gordo Trujillo G, López Meseguer M, Román Broto A, Lapunzina Abadía P, Escribano Subía P. An homozygous mutation in KCNK3 is associated with an aggressive form of hereditary pulmonary arterial hypertension. Clin Genet. 2017; 91:453–457.60. Dookeran KA, Zhang W, Stayner L, Argos M. Associations of two-pore domain potassium channels and triple negative breast cancer subtype in The Cancer Genome Atlas: systematic evaluation of gene expression and methylation. BMC Res Notes. 2017; 10:475.
Article61. Qu C, Sun J, Liu Y, Wang X, Wang L, Han C, Chen Q, Guan T, Li H, Zhang Y, Wang Y, Liu J, Zou W, Liu J. Caveolin-1 facilitated KCNA5 expression, promoting breast cancer viability. Oncol Lett. 2018; 16:4829–4838.
Article62. Bulk E, Ay AS, Hammadi M, Ouadid-Ahidouch H, Schelhaas S, Hascher A, Rohde C, Thoennissen NH, Wiewrodt R, Schmidt E, Marra A, Hillejan L, Jacobs AH, Klein HU, Dugas M, Berdel WE, Müller-Tidow C, Schwab A. Epigenetic dysregulation of KCa 31 channels induces poor prognosis in lung cancer. Int J Cancer. 2015; 137:1306–1317.63. Warnecke-Eberz U, Metzger R, Hölscher AH, Drebber U, Bollschweiler E. Diagnostic marker signature for esophageal cancer from transcriptome analysis. Tumour Biol. 2016; 37:6349–6358.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- K+ channel
- Expression of ATP-sensitive potassium channel and sulfonylurea receptor in neonate and adult rat tissues
- Change of voltage-gated potassium channel 1.7 expressions in monocrotaline-induced pulmonary arterial hypertension rat model
- The large-conductance calcium-activated potassium channel holds the key to the conundrum of familial hypokalemic periodic paralysis
- Systemic Administration of the Potassium Channel Activator in the Polystyrene Latex Bead-Induced Cerebral Vasospasm