Intest Res.
2019 Apr;17(2):177-191. 10.5217/ir.2018.00170.
Epithelial-microbial diplomacy: escalating border tensions drive inflammation in inflammatory bowel disease
- Affiliations
-
- 1Division of Biomedical Sciences, University of California, Riverside, CA, USA. declan.mccole@ucr.edu
- KMID: 2449959
- DOI: http://doi.org/10.5217/ir.2018.00170
Abstract
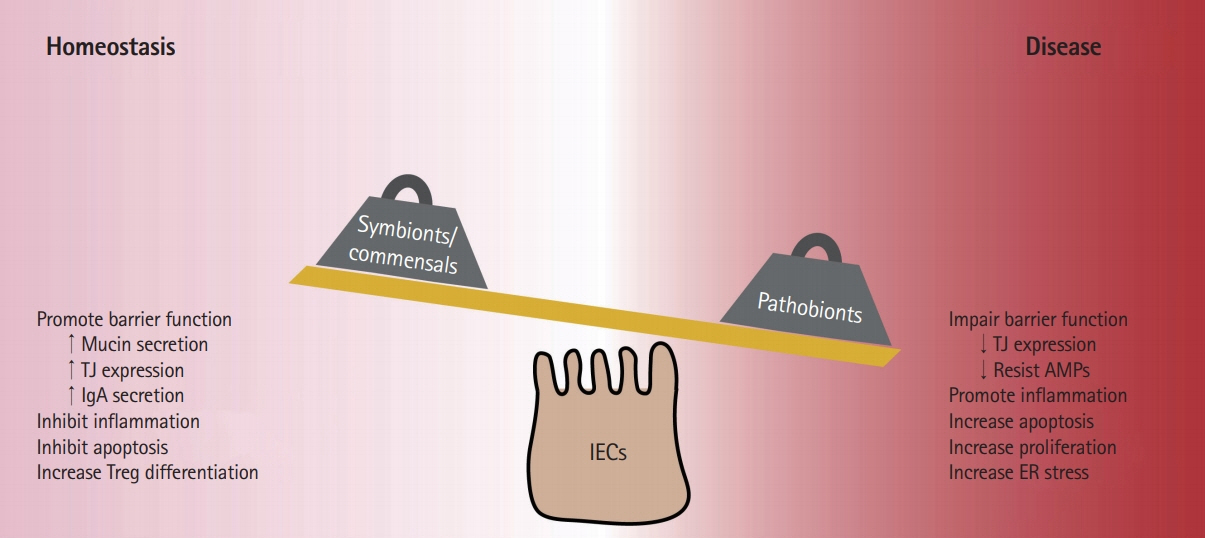
- Inflammatory bowel diseases (IBD) are chronic conditions of the gastrointestinal tract-the main site of host-microbial interaction in the body. Development of IBD is not due to a single event but rather is a multifactorial process where a patient's genetic background, behavioral habits, and environmental exposures contribute to disease pathogenesis. IBD patients exhibit alterations to gut bacterial populations "dysbiosis" due to the inflammatory microenvironment, however whether this alteration of the gut microbiota precedes inflammation has not been confirmed. Emerging evidence has highlighted the important role of gut microbes in developing measured immune responses and modulating other host responses such as metabolism. Much of the work on the gut microbiota has been correlative and there is an increasing need to understand the intimate relationship between host and microbe. In this review, we highlight how commensal and pathogenic bacteria interact with host intestinal epithelial cells and explore how altered microenvironments impact these connections.
MeSH Terms
Figure
Cited by 2 articles
-
Melatonin in the colon modulates intestinal microbiota in response to stress and sleep deprivation
Young Sook Park, Soo Hyung Kim, Jong Won Park, Younglim Kho, Pu Rum Seok, Jae-Ho Shin, Yoon Ji Choi, Jin-Hyun Jun, Hee Chan Jung, Eun Kyung Kim
Intest Res. 2020;18(3):325-336. doi: 10.5217/ir.2019.00093.Pathogenesis and clinical perspectives of extraintestinal manifestations in inflammatory bowel diseases
Jung Min Kim, Jae Hee Cheon
Intest Res. 2020;18(3):249-264. doi: 10.5217/ir.2019.00128.
Reference
-
1. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018; 390:2769–2778.
Article2. Dahlhamer JM, Zammitti EP, Ward BW, Wheaton AG, Croft JB. Prevalence of inflammatory bowel disease among adults aged ≥18 years - United States, 2015. MMWR Morb Mortal Wkly Rep. 2016; 65:1166–1169.
Article3. Centers for Disease Control and Prevention. Data and statistics: inflammatory bowel disease prevalence (IBD) in the United States. CDC Web site. (www.cdc.gov/ibd/data-statistics.htm. Updated November 14, 2018. Accessed February 13, 2019.4. Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis. 1999; 5:262–270.
Article5. Roulis M, Bongers G, Armaka M, et al. Host and microbiota interactions are critical for development of murine Crohn’slike ileitis. Mucosal Immunol. 2016; 9:787–797.
Article6. Khan KJ, Ullman TA, Ford AC, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and metaanalysis. Am J Gastroenterol. 2011; 106:661–673.
Article7. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533.
Article8. Davenport ER, Sanders JG, Song SJ, Amato KR, Clark AG, Knight R. The human microbiome in evolution. BMC Biol. 2017; 15:127.
Article9. Maier E, Anderson RC, Roy NC. Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients. 2014; 7:45–73.
Article10. Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol. 2015; 6:639.
Article11. Lee N, Kim WU. Microbiota in T-cell homeostasis and inflammatory diseases. Exp Mol Med. 2017; 49:e340.
Article12. Verma R, Lee C, Jeun EJ, et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci Immunol. 2018; 3:eaat6975.
Article13. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–1031.
Article14. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007; 104:13780–13785.
Article15. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017; 17:219–232.
Article16. Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013; 14:660–667.
Article17. Watson AJ, Duckworth CA, Guan Y, Montrose MH. Mechanisms of epithelial cell shedding in the mammalian intestine and maintenance of barrier function. Ann N Y Acad Sci. 2009; 1165:135–142.
Article18. Marchiando AM, Shen L, Graham WV, et al. The epithelial barrier is maintained by in vivo tight junction expansion during pathologic intestinal epithelial shedding. Gastroenterology. 2011; 140:1208–1218.
Article19. Blander JM. Death in the intestinal epithelium-basic biology and implications for inflammatory bowel disease. FEBS J. 2016; 283:2720–2730.
Article20. Tsai PY, Zhang B, He WQ, et al. IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe. 2017; 21:671–681.21. Chairatana P, Nolan EM. Defensins, lectins, mucins, and secretory immunoglobulin A: microbe-binding biomolecules that contribute to mucosal immunity in the human gut. Crit Rev Biochem Mol Biol. 2017; 52:45–56.
Article22. Ermund A, Schütte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013; 305:G341–G347.
Article23. Johansson ME, Jakobsson HE, Holmén-Larsson J, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015; 18:582–592.
Article24. Van Tassell ML, Miller MJ. Lactobacillus adhesion to mucus. Nutrients. 2011; 3:613–636.
Article25. Norkina O, Burnett TG, De Lisle RC. Bacterial overgrowth in the cystic fibrosis transmembrane conductance regulator null mouse small intestine. Infect Immun. 2004; 72:6040–6049.
Article26. Bazett M, Honeyman L, Stefanov AN, Pope CE, Hoffman LR, Haston CK. Cystic fibrosis mouse model-dependent intestinal structure and gut microbiome. Mamm Genome. 2015; 26:222–234.
Article27. Price AE, Shamardani K, Lugo KA, et al. A map of Toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018; 49:560–575.
Article28. Vora P, Youdim A, Thomas LS, et al. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol. 2004; 173:5398–5405.
Article29. Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007; 132:1359–1374.
Article30. Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011; 141:769–776.
Article31. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009; 139:485–498.
Article32. Goto Y, Panea C, Nakato G, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014; 40:594–607.
Article33. Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015; 163:367–380.
Article34. Sano T, Huang W, Hall JA, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell. 2015; 163:381–393.
Article35. Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A. 2016; 113:E8141–E8150.
Article36. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005; 122:107–118.
Article37. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008; 453:620–625.
Article38. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory Tcell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010; 107:12204–12209.
Article39. Knoop KA, Newberry RD. Isolated lymphoid follicles are dynamic reservoirs for the induction of intestinal IgA. Front Immunol. 2012; 3:84.
Article40. Wu W, Sun M, Chen F, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017; 10:946–956.
Article41. Wang Y, Liu L, Moore DJ, et al. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol. 2017; 10:373–384.
Article42. Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003; 109:580–587.
Article43. Mathias A, Duc M, Favre L, Benyacoub J, Blum S, Corthésy B. Potentiation of polarized intestinal Caco-2 cell responsiveness to probiotics complexed with secretory IgA. J Biol Chem. 2010; 285:33906–33913.
Article44. Donaldson GP, Ladinsky MS, Yu KB, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018; 360:795–800.
Article45. Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014; 158:1000–1010.
Article46. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001; 411:599–603.
Article47. Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008; 40:955–962.48. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010; 42:1118–1125.49. McGovern DP, Gardet A, Törkvist L, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010; 42:332–337.50. Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011; 43:246–252.51. Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009; 106:15813–15818.
Article52. Rehman A, Sina C, Gavrilova O, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011; 60:1354–1362.
Article53. Mondot S, Barreau F, Al Nabhani Z, et al. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut. 2012; 61:634–635.
Article54. Sadaghian Sadabad M, Regeling A, de Goffau MC, et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn’s disease patients. Gut. 2015; 64:1546–1552.
Article55. Pott J, Kabat AM, Maloy KJ. Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe. 2018; 23:191–202.
Article56. McCole DF. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. 2014; 20:1829–1849.
Article57. Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014; 6:107.
Article58. Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013; 502:96–99.
Article59. Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003; 299:2074–2076.
Article60. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucindegrading bacterium. Int J Syst Evol Microbiol. 2004; 54:1469–1476.
Article61. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005; 307:1915–1920.
Article62. Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010; 105:2420–2428.
Article63. van Passel MW, Kant R, Zoetendal EG, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. 2011; 6:e16876.
Article64. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010; 10:159–169.
Article65. Van den Abbeele P, Belzer C, Goossens M, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013; 7:949–961.
Article66. Robinson LS, Lewis WG, Lewis AL. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidasemediated cross-species foraging of 9-O-acetylated sialoglycans. J Biol Chem. 2017; 292:11861–11872.
Article67. Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. 2012; 78:420–428.
Article68. Belzer C, Chia LW, Aalvink S, et al. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B(12) production by intestinal symbionts. MBio. 2017; 85:e00770–17.69. Berry D, Stecher B, Schintlmeister A, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A. 2013; 110:4720–4725.
Article70. Reunanen J, Kainulainen V, Huuskonen L, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. 2015; 81:3655–3662.
Article71. Ouwerkerk JP, van der Ark KCH, Davids M, et al. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl Environ Microbiol. 2016; 82:6983–6993.
Article72. Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013; 110:9066–9071.
Article73. Shin NR, Lee JC, Lee HY, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014; 63:727–735.
Article74. Ottman N, Reunanen J, Meijerink M, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017; 12:e0173004.
Article75. Gaudier E, Jarry A, Blottière HM, et al. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol. 2004; 287:G1168–G1174.76. Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis. 2010; 16:684–695.
Article77. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016; 165:1332–1345.
Article78. Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018; 362:eaat9076.
Article79. Alam A, Leoni G, Quiros M, et al. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016; 1:15021.
Article80. Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013; 19:481–488.
Article81. Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011; 60:34–40.
Article82. Karlsson CL, Onnerfält J, Xu J, Molin G, Ahrné S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring). 2012; 20:2257–2261.
Article83. Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016; 65:426–436.
Article84. Zhang YG, Wu S, Xia Y, Sun J. Salmonella infection upregulates the leaky protein claudin-2 in intestinal epithelial cells. PLoS One. 2013; 8:e58606.
Article85. Chelakkot C, Choi Y, Kim DK, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med. 2018; 50:e450.
Article86. Wlodarska M, Luo C, Kolde R, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017; 22:25–37.
Article87. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009; 15:1183–1189.
Article88. Lopez-Siles M, Martinez-Medina M, Abellà C, et al. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol. 2015; 81:7582–7592.
Article89. Lopez-Siles M, Enrich-Capó N, Aldeguer X, et al. Alterations in the abundance and co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the colonic mucosa of inflammatory bowel disease subjects. Front Cell Infect Microbiol. 2018; 8:281.
Article90. Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180.
Article91. Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract. 2014; 2014:872725.92. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008; 105:16731–16736.
Article93. Jia W, Whitehead RN, Griffiths L, et al. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn’s disease? FEMS Microbiol Lett. 2010; 310:138–144.
Article94. Quévrain E, Maubert MA, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016; 65:415–425.
Article95. Breyner NM, Michon C, de Sousa CS, et al. Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-kappaB pathway. Front Microbiol. 2017; 8:114.96. Maier E, Anderson RC, Roy NC. Live Faecalibacterium prausnitzii does not enhance epithelial barrier integrity in an apical anaerobic co-culture model of the large intestine. Nutrients. 2017; 9:1349.
Article97. He X, Zeng Q, Puthiyakunnon S, et al. Lactobacillus rhamnosus GG supernatant enhance neonatal resistance to systemic Escherichia coli K1 infection by accelerating development of intestinal defense. Sci Rep. 2017; 7:43305.
Article98. Alam A, Leoni G, Wentworth CC, et al. Redox signaling regulates commensal-mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunol. 2014; 7:645–655.
Article99. Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007; 132:562–575.
Article100. Shen X, Liu L, Peek RM, et al. Supplementation of p40, a Lactobacillus rhamnosus GG-derived protein, in early life promotes epidermal growth factor receptor-dependent intestinal development and long-term health outcomes. Mucosal Immunol. 2018; 11:1316–1328.
Article101. Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004; 427:441–444.
Article102. Tajkarimi M, Wexler HM. CRISPR-Cas systems in Bacteroides fragilis, an important pathobiont in the human gut microbiome. Front Microbiol. 2017; 8:2234.
Article103. Chan JL, Wu S, Geis AL, et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019; 12:164–177.
Article104. Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006; 44:3980–3988.
Article105. Patwa LG, Fan TJ, Tchaptchet S, et al. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011; 141:1842–1851.
Article106. Tchaptchet S, Fan TJ, Goeser L, et al. Inflammation-induced acid tolerance genes gadAB in luminal commensal Escherichia coli attenuate experimental colitis. Infect Immun. 2013; 81:3662–3671.
Article107. Ocvirk S, Sava IG, Lengfelder I, et al. Surface-associated lipoproteins link Enterococcus faecalis virulence to colitogenic activity in IL-10-deficient mice independent of their expression levels. PLoS Pathog. 2015; 11:e1004911.
Article108. Chassaing B, Gewirtz AT. Mice harboring pathobiont-free microbiota do not develop intestinal inflammation that normally results from an innate immune deficiency. PLoS One. 2018; 13:e0195310.
Article109. Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014; 124:3617–3633.
Article110. Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008; 135:568–579.
Article111. Medani M, Collins D, Docherty NG, Baird AW, O’Connell PR, Winter DC. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm Bowel Dis. 2011; 17:1620–1625.
Article112. Thiennimitr P, Winter SE, Winter MG, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011; 108:17480–17485.
Article113. Pham TA, Clare S, Goulding D, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014; 16:504–516.
Article114. Zhu W, Winter MG, Byndloss MX, et al. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018; 553:208–211.
Article115. Deriu E, Liu JZ, Pezeshki M, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013; 14:26–37.
Article116. Bolick DT, Kolling GL, Moore JH 2nd, et al. Zinc deficiency alters host response and pathogen virulence in a mouse model of enteroaggregative Escherichia coli-induced diarrhea. Gut Microbes. 2014; 5:618–627.
Article117. Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012; 487:104–108.
Article118. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019; 4:293–305.
Article119. Albenberg L, Esipova TV, Judge CP, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014; 147:1055–1063.
Article120. Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007; 2:204.
Article121. Rivera-Chávez F, Zhang LF, Faber F, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016; 19:443–454.
Article122. Fiorucci S, Distrutti E, Cirino G, Wallace JL. The emerging roles of hydrogen sulfide in the gastrointestinal tract and liver. Gastroenterology. 2006; 131:259–271.
Article123. Bertin Y, Girardeau JP, Chaucheyras-Durand F, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011; 13:365–377.
Article124. Faber F, Thiennimitr P, Spiga L, et al. Respiration of microbiota-derived 1,2-propanediol zdrives Salmonella expansion during colitis. PLoS Pathog. 2017; 13:e1006129.125. Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009; 461:1282–1286.
Article126. Hughes ER, Winter MG, Duerkop BA, et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe. 2017; 21:208–219.
Article127. Lopez CA, Skaar EP. The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe. 2018; 23:737–748.
Article128. Blachier F, Beaumont M, Andriamihaja M, et al. Changes in the luminal environment of the colonic epithelial cells and physiopathological consequences. Am J Pathol. 2017; 187:476–486.
Article129. Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005; 71:3692–3700.
Article130. Talbot C, Lytle C. Segregation of Na/H exchanger-3 and Cl/HCO3 exchanger SLC26A3 (DRA) in rodent cecum and colon. Am J Physiol Gastrointest Liver Physiol. 2010; 299:G358–G367.131. Vernia P, Caprilli R, Latella G, Barbetti F, Magliocca FM, Cittadini M. Fecal lactate and ulcerative colitis. Gastroenterology. 1988; 95:1564–1568.
Article132. Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol. 2007; 73:6526–6533.
Article133. Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010; 7:265–276.
Article134. Wu C, Sartor RB, Huang K, Tonkonogy SL. Transient activation of mucosal effector immune responses by resident intestinal bacteria in normal hosts is regulated by interleukin-10 signalling. Immunology. 2016; 148:304–314.
Article135. Gomes-Neto JC, Kittana H, Mantz S, et al. A gut pathobiont synergizes with the microbiota to instigate inflammatory disease marked by immunoreactivity against other symbionts but not itself. Sci Rep. 2017; 7:17707.
Article136. Bretin A, Lucas C, Larabi A, et al. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci Rep. 2018; 8:12301.
Article137. Wells CL, Jechorek RP, Erlandsen SL. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J Infect Dis. 1990; 162:82–90.
Article138. Runkel NS, Moody FG, Smith GS, et al. Alterations in rat intestinal transit by morphine promote bacterial translocation. Dig Dis Sci. 1993; 38:1530–1536.
Article139. Vinderola CG, Medici M, Perdigón G. Relationship between interaction sites in the gut, hydrophobicity, mucosal immunomodulating capacities and cell wall protein profiles in indigenous and exogenous bacteria. J Appl Microbiol. 2004; 96:230–243.
Article140. Gerardo SH, Garcia MM, Wexler HM, Finegold SM. Adherence of Bilophila wadsworthia to cultured human embryonic intestinal cells. Anaerobe. 1998; 4:19–27.
Article141. Zeng J, Teng F, Murray BE. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect Immun. 2005; 73:1606–1612.
Article142. Zeng J, Teng F, Weinstock GM, Murray BE. Translocation of Enterococcus faecalis strains across a monolayer of polarized human enterocyte-like T84 cells. J Clin Microbiol. 2004; 42:1149–1154.
Article143. Vazeille E, Chassaing B, Buisson A, et al. GipA factor supports colonization of Peyer’s patches by Crohn’s disease-associated Escherichia coli. Inflamm Bowel Dis. 2016; 22:68–81.
Article144. Zhang L, Man SM, Day AS, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J Clin Microbiol. 2009; 47:453–455.
Article145. Mahendran V, Riordan SM, Grimm MC, et al. Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS One. 2011; 6:e25417.
Article146. Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One. 2011; 6:e21490.
Article147. Man SM, Kaakoush NO, Leach ST, et al. Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J Infect Dis. 2010; 202:1855–1865.
Article148. Nielsen HL, Nielsen H, Ejlertsen T, et al. Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. PLoS One. 2011; 6:e23858.
Article149. Raisch J, Buc E, Bonnet M, et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol. 2014; 20:6560–6572.
Article150. Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014; 63:1932–1942.
Article151. Arthur JC, Jobin C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes. 2013; 4:253–258.152. Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014; 41:311–324.
Article153. Charlet R, Pruvost Y, Tumba G, et al. Remodeling of the Candida glabrata cell wall in the gastrointestinal tract affects the gut microbiota and the immune response. Sci Rep. 2018; 8:3316.
Article154. Hoffmann M, Messlik A, Kim SC, Sartor RB, Haller D. Impact of a probiotic Enterococcus faecalis in a gnotobiotic mouse model of experimental colitis. Mol Nutr Food Res. 2011; 55:703–713.
Article155. Rolhion N, Barnich N, Bringer MA, et al. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010; 59:1355–1362.
Article156. Rolhion N, Hofman P, Darfeuille-Michaud A. The endoplasmic reticulum stress response chaperone: Gp96, a host receptor for Crohn disease-associated adherent-invasive Escherichia coli. Gut Microbes. 2011; 2:115–119.
Article157. Clayton JB, Vangay P, Huang H, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci U S A. 2016; 113:10376–10381.
Article158. Banla IL, Kommineni S, Hayward M, et al. Modulators of Enterococcus faecalis cell envelope integrity and antimicrobial resistance influence stable colonization of the mammalian gastrointestinal tract. Infect Immun. 2017; 86:e00381–17.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Can vitamin D supplementation help control inflammation in inflammatory bowel disease beyond its classical role in bone health?
- Intestinal microbiota and inflammatory bowel diseases
- Inflammatory Bowel Diseases and Enteric Microbiota
- Navigating the Microbial Basis of Inflammatory Bowel Diseases: Seeing the Light at the End of the Tunnel
- Antimicrobial Proteins in Intestine and Inflammatory Bowel Diseases