Endocrinol Metab.
2018 Sep;33(3):318-330. 10.3803/EnM.2018.33.3.318.
Regulation of Osteoblast Metabolism by Wnt Signaling
- Affiliations
-
- 1Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, MD, USA. rriddle1@jhmi.edu
- 2Baltimore Veterans Administration Medical Center, Baltimore, MD, USA.
- KMID: 2447021
- DOI: http://doi.org/10.3803/EnM.2018.33.3.318
Abstract
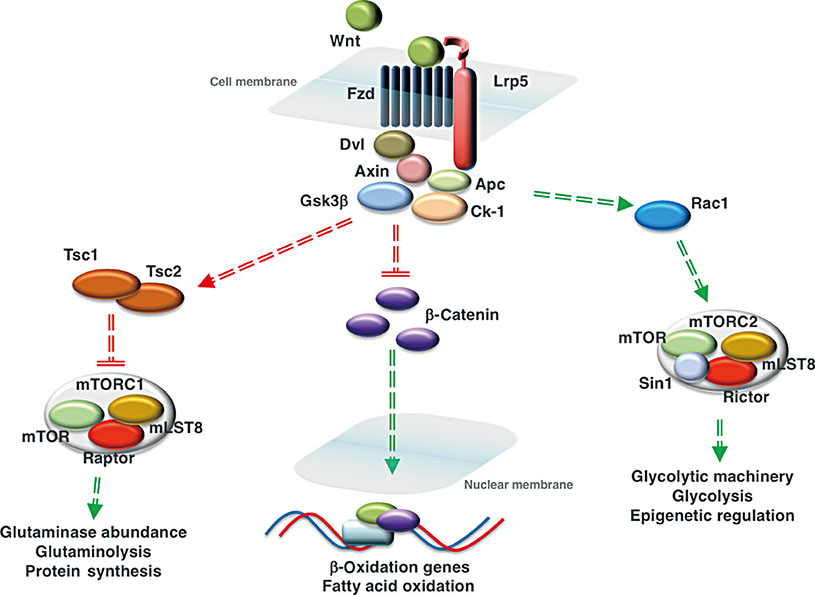
- Wnt/β-catenin signaling plays a critical role in the achievement of peak bone mass, affecting the commitment of mesenchymal progenitors to the osteoblast lineage and the anabolic capacity of osteoblasts depositing bone matrix. Recent studies suggest that this evolutionarily-conserved, developmental pathway exerts its anabolic effects in part by coordinating osteoblast activity with intermediary metabolism. These findings are compatible with the cloning of the gene encoding the low-density lipoprotein related receptor-5 (LRP5) Wnt co-receptor from a diabetes-susceptibility locus and the now well-established linkage between Wnt signaling and metabolism. In this article, we provide an overview of the role of Wnt signaling in whole-body metabolism and review the literature regarding the impact of Wnt signaling on the osteoblast's utilization of three different energy sources: fatty acids, glucose, and glutamine. Special attention is devoted to the net effect of nutrient utilization and the mode of regulation by Wnt signaling. Mechanistic studies indicate that the utilization of each substrate is governed by a unique mechanism of control with β-catenin-dependent signaling regulating fatty acid β-oxidation, while glucose and glutamine utilization are β-catenin-independent and downstream of mammalian target of rapamycin complex 2 (mTORC2) and mammalian target of rapamycin complex 1 (mTORC1) activation, respectively. The emergence of these data has provided a new context for the mechanisms by which Wnt signaling influences bone development.
MeSH Terms
Figure
Cited by 1 articles
-
Changes in Serum Dickkopf-1, RANK Ligand, Osteoprotegerin, and Bone Mineral Density after Allogeneic Hematopoietic Stem Cell Transplantation Treatment
Eunhee Jang, Jeonghoon Ha, Ki-Hyun Baek, Moo Il Kang
Endocrinol Metab. 2021;36(6):1211-1218. doi: 10.3803/EnM.2021.1248.
Reference
-
1. Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013; 5:a008334.
Article2. Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011; 13:27–38.
Article3. Neuman MW, Neuman WF. Emerging concepts of the structure and metabolic functions of bone. Am J Med. 1957; 22:123–131.
Article4. Neuman WF, Neuman MW, Brommage R. Aerobic glycolysis in bone: lactate production and gradients in calvaria. Am J Physiol. 1978; 234:C41–C50.
Article5. Borle AB, Nichols N, Nichols G Jr. Metabolic studies of bone in vitro. II. The metabolic patterns of accretion and resorption. J Biol Chem. 1960; 235:1211–1214.6. Borle AB, Nichols N, Nichols G Jr. Metabolic studies of bone in vitro. I. Normal bone. J Biol Chem. 1960; 235:1206–1210.7. Flanagan B, Nichols G Jr. Metabolic studies of bone in vitro. V. Glucose metabolism and collagen biosynthesis. J Biol Chem. 1964; 239:1261–1265.8. Adamek G, Felix R, Guenther HL, Fleisch H. Fatty acid oxidation in bone tissue and bone cells in culture. Characterization and hormonal influences. Biochem J. 1987; 248:129–137.
Article9. Guntur AR, Le PT, Farber CR, Rosen CJ. Bioenergetics during calvarial osteoblast differentiation reflect strain differences in bone mass. Endocrinology. 2014; 155:1589–1595.
Article10. Komarova SV, Ataullakhanov FI, Globus RK. Bioenergetics and mitochondrial transmembrane potential during differentiation of cultured osteoblasts. Am J Physiol Cell Physiol. 2000; 279:C1220–C1229.
Article11. Wei J, Shimazu J, Makinistoglu MP, Maurizi A, Kajimura D, Zong H, et al. Glucose uptake and runx2 synergize to orchestrate osteoblast differentiation and bone formation. Cell. 2015; 161:1576–1591.
Article12. Niemeier A, Niedzielska D, Secer R, Schilling A, Merkel M, Enrich C, et al. Uptake of postprandial lipoproteins into bone in vivo: impact on osteoblast function. Bone. 2008; 43:230–237.
Article13. Kim SP, Li Z, Zoch ML, Frey JL, Bowman CE, Kushwaha P, et al. Fatty acid oxidation by the osteoblast is required for normal bone acquisition in a sex- and diet-dependent manner. JCI Insight. 2017; 2:e92704.
Article14. Fulzele K, Riddle RC, DiGirolamo DJ, Cao X, Wan C, Chen D, et al. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell. 2010; 142:309–319.
Article15. Li Z, Frey JL, Wong GW, Faugere MC, Wolfgang MJ, Kim JK, et al. Glucose transporter-4 facilitates insulin-stimulated glucose uptake in osteoblasts. Endocrinology. 2016; 157:4094–4103.
Article16. Dirckx N, Tower RJ, Mercken EM, Vangoitsenhoven R, Moreau-Triby C, Breugelmans T, et al. Vhl deletion in osteoblasts boosts cellular glycolysis and improves global glucose metabolism. J Clin Invest. 2018; 128:1087–1105.
Article17. Stegen S, van Gastel N, Eelen G, Ghesquiere B, D'Anna F, Thienpont B, et al. HIF-1α promotes glutamine-mediated redox homeostasis and glycogen-dependent bioenergetics to support postimplantation bone cell survival. Cell Metab. 2016; 23:265–279.
Article18. Regan JN, Lim J, Shi Y, Joeng KS, Arbeit JM, Shohet RV, et al. Up-regulation of glycolytic metabolism is required for HIF1α-driven bone formation. Proc Natl Acad Sci U S A. 2014; 111:8673–8678.
Article19. Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012; 149:1192–1205.
Article20. Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009; 10:468–477.
Article21. Jin T. The WNT signalling pathway and diabetes mellitus. Diabetologia. 2008; 51:1771–1780.
Article22. Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, et al. Association of the APC gene product with beta-catenin. Science. 1993; 262:1731–1734.
Article23. Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007; 316:1043–1046.24. Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998; 17:1371–1384.25. Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, et al. Axin, a negative regulator of the Wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta-catenin. J Biol Chem. 1998; 273:10823–10826.26. Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002; 108:837–847.27. Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997; 16:3797–3804.28. Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, et al. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999; 18:2401–2410.29. Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012; 149:1245–1256.
Article30. He X, Saint-Jeannet JP, Wang Y, Nathans J, Dawid I, Varmus H. A member of the Frizzled protein family mediating axis induction by Wnt-5A. Science. 1997; 275:1652–1654.
Article31. Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, et al. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996; 382:225–230.
Article32. Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, et al. Arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000; 407:527–530.
Article33. Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000; 407:530–535.
Article34. Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005; 438:873–877.
Article35. Smalley MJ, Sara E, Paterson H, Naylor S, Cook D, Jayatilake H, et al. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO J. 1999; 18:2823–2835.
Article36. Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A. DIX domains of Dvl and axin are necessary for protein interactions and their ability to regulate beta-catenin stability. Mol Cell Biol. 1999; 19:4414–4422.37. Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, et al. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development. 2008; 135:367–375.
Article38. Mao J, Wang J, Liu B, Pan W, Farr GH 3rd, Flynn C, et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001; 7:801–809.
Article39. Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996; 86:391–399.40. Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996; 382:638–642.41. Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, et al. Drosophila Tcf and Groucho interact to repress Wingless signaling activity. Nature. 1998; 395:604–608.42. Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998; 395:608–612.
Article43. Lee W, Swarup S, Chen J, Ishitani T, Verheyen EM. Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development. 2009; 136:241–251.44. Hikasa H, Ezan J, Itoh K, Li X, Klymkowsky MW, Sokol SY. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev Cell. 2010; 19:521–532.
Article45. Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000; 19:1839–1850.
Article46. Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000; 149:249–254.47. Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001; 20:4935–4943.
Article48. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003; 115:577–590.
Article49. Valvezan AJ, Huang J, Lengner CJ, Pack M, Klein PS. Oncogenic mutations in adenomatous polyposis coli (Apc) activate mechanistic target of rapamycin complex 1 (mTORC1) in mice and zebrafish. Dis Model Mech. 2014; 7:63–71.50. Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, Long F. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 2013; 17:745–755.
Article51. Sun W, Shi Y, Lee WC, Lee SY, Long F. Rictor is required for optimal bone accrual in response to anti-sclerostin therapy in the mouse. Bone. 2016; 85:1–8.
Article52. van Amerongen R. Alternative Wnt pathways and receptors. Cold Spring Harb Perspect Biol. 2012; 4:a007914.
Article53. Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005; 38:439–446.54. Yang Y, Mlodzik M. Wnt-Frizzled/planar cell polarity signaling: cellular orientation by facing the wind (Wnt). Annu Rev Cell Dev Biol. 2015; 31:623–646.
Article55. Torres MA, Yang-Snyder JA, Purcell SM, DeMarais AA, McGrew LL, Moon RT. Activities of the Wnt-1 class of secreted signaling factors are antagonized by the Wnt-5A class and by a dominant negative cadherin in early Xenopus development. J Cell Biol. 1996; 133:1123–1137.
Article56. Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007; 134:479–489.
Article57. Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J Cell Biol. 2003; 162:899–908.58. Westfall TA, Brimeyer R, Twedt J, Gladon J, Olberding A, Furutani-Seiki M, et al. Wnt-5/pipetail functions in vertebrate axis formation as a negative regulator of Wnt/beta-catenin activity. J Cell Biol. 2003; 162:889–898.59. Slusarski DC, Corces VG, Moon RT. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997; 390:410–413.
Article60. Slusarski DC, Yang-Snyder J, Busa WB, Moon RT. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol. 1997; 182:114–120.61. Kuhl M, Sheldahl LC, Malbon CC, Moon RT. Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J Biol Chem. 2000; 275:12701–12711.62. Sheldahl LC, Park M, Malbon CC, Moon RT. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr Biol. 1999; 9:695–698.63. Gao B, Yang Y. Planar cell polarity in vertebrate limb morphogenesis. Curr Opin Genet Dev. 2013; 23:438–444.
Article64. Andre P, Song H, Kim W, Kispert A, Yang Y. Wnt5a and Wnt11 regulate mammalian anterior-posterior axis elongation. Development. 2015; 142:1516–1527.
Article65. Gros J, Serralbo O, Marcelle C. WNT11 acts as a directional cue to organize the elongation of early muscle fibres. Nature. 2009; 457:589–593.
Article66. Tada M, Smith JC. Xwnt11 is a target of Xenopus Brachyury: regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development. 2000; 127:2227–2238.
Article67. Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994; 8:174–189.
Article68. Greco TL, Takada S, Newhouse MM, McMahon JA, McMahon AP, Camper SA. Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev. 1996; 10:313–324.
Article69. Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001; 107:513–523.70. Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002; 70:11–19.
Article71. Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002; 346:1513–1521.
Article72. Ellies DL, Viviano B, McCarthy J, Rey JP, Itasaki N, Saunders S, et al. Bone density ligand, sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006; 21:1738–1749.
Article73. Semenov MV, He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006; 281:38276–38284.
Article74. Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005; 25:4946–4955.
Article75. Bhat BM, Allen KM, Liu W, Graham J, Morales A, Anisowicz A, et al. Structure-based mutation analysis shows the importance of LRP5 beta-propeller 1 in modulating Dkk1-mediated inhibition of Wnt signaling. Gene. 2007; 391:103–112.76. Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, Grover M, et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med. 2013; 368:1809–1816.
Article77. Garcia-Ibarbia C, Perez-Nunez MI, Olmos JM, Valero C, Perez-Aguilar MD, Hernandez JL, et al. Missense polymorphisms of the WNT16 gene are associated with bone mass, hip geometry and fractures. Osteoporos Int. 2013; 24:2449–2454.
Article78. Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, Nelson-Williams C, et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007; 315:1278–1282.
Article79. Van Wesenbeeck L, Cleiren E, Gram J, Beals RK, Benichou O, Scopelliti D, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003; 72:763–771.
Article80. Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002; 39:91–97.
Article81. Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA 2nd, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002; 157:303–314.
Article82. Riddle RC, Diegel CR, Leslie JM, Van Koevering KK, Faugere MC, Clemens TL, et al. Lrp5 and Lrp6 exert overlapping functions in osteoblasts during postnatal bone acquisition. PLoS One. 2013; 8:e63323.
Article83. Babij P, Zhao W, Small C, Kharode Y, Yaworsky PJ, Bouxsein ML, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003; 18:960–974.
Article84. Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005; 8:727–738.85. Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005; 8:739–750.86. Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF, et al. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res. 2004; 19:2033–2040.
Article87. Zhong Z, Zylstra-Diegel CR, Schumacher CA, Baker JJ, Carpenter AC, Rao S, et al. Wntless functions in mature osteoblasts to regulate bone mass. Proc Natl Acad Sci U S A. 2012; 109:E2197–E2204.
Article88. Glass DA 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005; 8:751–764.
Article89. Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, et al. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem. 2005; 280:21162–21168.90. Li C, Xing Q, Yu B, Xie H, Wang W, Shi C, et al. Disruption of LRP6 in osteoblasts blunts the bone anabolic activity of PTH. J Bone Miner Res. 2013; 28:2094–2108.
Article91. Romero G, Sneddon WB, Yang Y, Wheeler D, Blair HC, Friedman PA. Parathyroid hormone receptor directly interacts with dishevelled to regulate beta-catenin signaling and osteoclastogenesis. J Biol Chem. 2010; 285:14756–14763.92. Wan M, Yang C, Li J, Wu X, Yuan H, Ma H, et al. Parathyroid hormone signaling through low-density lipoprotein-related protein 6. Genes Dev. 2008; 22:2968–2979.
Article93. Kulkarni NH, Halladay DL, Miles RR, Gilbert LM, Frolik CA, Galvin RJ, et al. Effects of parathyroid hormone on Wnt signaling pathway in bone. J Cell Biochem. 2005; 95:1178–1190.
Article94. Kramer I, Halleux C, Keller H, Pegurri M, Gooi JH, Weber PB, et al. Osteocyte Wnt/beta-catenin signaling is required for normal bone homeostasis. Mol Cell Biol. 2010; 30:3071–3085.95. Twells RC, Metzker ML, Brown SD, Cox R, Garey C, Hammond H, et al. The sequence and gene characterization of a 400-kb candidate region for IDDM4 on chromosome 11q13. Genomics. 2001; 72:231–242.
Article96. Hey PJ, Twells RC, Phillips MS, Nakagawa Y, Brown SD, Kawaguchi Y, et al. Cloning of a novel member of the low-density lipoprotein receptor family. Gene. 1998; 216:103–111.
Article97. Twells RC, Mein CA, Payne F, Veijola R, Gilbey M, Bright M, et al. Linkage and association mapping of the LRP5 locus on chromosome 11q13 in type 1 diabetes. Hum Genet. 2003; 113:99–105.98. Suwazono Y, Kobayashi E, Uetani M, Miura K, Morikawa Y, Ishizaki M, et al. G-protein beta 3 subunit polymorphism C1429T and low-density lipoprotein receptor-related protein 5 polymorphism A1330V are risk factors for hypercholesterolemia in Japanese males: a prospective study over 5 years. Metabolism. 2006; 55:751–757.99. Suwazono Y, Kobayashi E, Uetani M, Miura K, Morikawa Y, Ishizaki M, et al. Low-density lipoprotein receptor-related protein 5 variant Q89R is associated with hypertension in Japanese females. Blood Press. 2006; 15:80–87.
Article100. Suwazono Y, Kobayashi E, Dochi M, Miura K, Morikawa Y, Ishizaki M, et al. Combination of the C1429T polymorphism in the G-protein beta-3 subunit gene and the A1330V polymorphism in the low-density lipoprotein receptor-related protein 5 gene is a risk factor for hypercholesterolaemia. Clin Exp Med. 2007; 7:108–114.
Article101. Guo YF, Xiong DH, Shen H, Zhao LJ, Xiao P, Guo Y, et al. Polymorphisms of the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with obesity phenotypes in a large family-based association study. J Med Genet. 2006; 43:798–803.
Article102. Lappalainen S, Saarinen A, Utriainen P, Voutilainen R, Jaaskelainen J, Makitie O. LRP5 in premature adrenarche and in metabolic characteristics of prepubertal children. Clin Endocrinol (Oxf). 2009; 70:725–731.103. Loh NY, Neville MJ, Marinou K, Hardcastle SA, Fielding BA, Duncan EL, et al. LRP5 regulates human body fat distribution by modulating adipose progenitor biology in a dose- and depot-specific fashion. Cell Metab. 2015; 21:262–273.
Article104. Kim DH, Inagaki Y, Suzuki T, Ioka RX, Yoshioka SZ, Magoori K, et al. A new low density lipoprotein receptor related protein, LRP5, is expressed in hepatocytes and adrenal cortex, and recognizes apolipoprotein E. J Biochem. 1998; 124:1072–1076.
Article105. Fujino T, Asaba H, Kang MJ, Ikeda Y, Sone H, Takada S, et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci U S A. 2003; 100:229–234.106. Magoori K, Kang MJ, Ito MR, Kakuuchi H, Ioka RX, Kamataki A, et al. Severe hypercholesterolemia, impaired fat tolerance, and advanced atherosclerosis in mice lacking both low density lipoprotein receptor-related protein 5 and apolipoprotein E. J Biol Chem. 2003; 278:11331–11336.
Article107. Kanazawa A, Tsukada S, Sekine A, Tsunoda T, Takahashi A, Kashiwagi A, et al. Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am J Hum Genet. 2004; 75:832–843.
Article108. Christodoulides C, Scarda A, Granzotto M, Milan G, Dalla Nora E, Keogh J, et al. WNT10B mutations in human obesity. Diabetologia. 2006; 49:678–684.
Article109. Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006; 38:320–323.
Article110. Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, et al. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006; 355:241–250.111. Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjogren M, et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in non-diabetic individuals. Diabetes. 2006; 55:2890–2895.
Article112. Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, Weitgasser R, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med (Berl). 2007; 85:777–782.
Article113. Columbus J, Chiang Y, Shao W, Zhang N, Wang D, Gaisano HY, et al. Insulin treatment and high-fat diet feeding reduces the expression of three Tcf genes in rodent pancreas. J Endocrinol. 2010; 207:77–86.
Article114. Ip W, Shao W, Chiang YT, Jin T. The Wnt signaling pathway effector TCF7L2 is upregulated by insulin and represses hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2012; 303:E1166–E1176.
Article115. Kaminska D, Kuulasmaa T, Venesmaa S, Kakela P, Vaittinen M, Pulkkinen L, et al. Adipose tissue TCF7L2 splicing is regulated by weight loss and associates with glucose and fatty acid metabolism. Diabetes. 2012; 61:2807–2813.
Article116. Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998; 19:379–383.
Article117. Savic D, Ye H, Aneas I, Park SY, Bell GI, Nobrega MA. Alterations in TCF7L2 expression define its role as a key regulator of glucose metabolism. Genome Res. 2011; 21:1417–1425.
Article118. da Silva Xavier G, Mondragon A, Sun G, Chen L, McGinty JA, French PM, et al. Abnormal glucose tolerance and insulin secretion in pancreas-specific Tcf7l2-null mice. Diabetologia. 2012; 55:2667–2676.
Article119. Mitchell RK, Mondragon A, Chen L, Mcginty JA, French PM, Ferrer J, et al. Selective disruption of Tcf7l2 in the pancreatic β cell impairs secretory function and lowers β cell mass. Hum Mol Genet. 2015; 24:1390–1399.
Article120. Boj SF, van Es JH, Huch M, Li VS, Jose A, Hatzis P, et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012; 151:1595–1607.
Article121. Jin T. Current understanding on role of the Wnt signaling pathway effector TCF7L2 in glucose homeostasis. Endocr Rev. 2016; 37:254–277.
Article122. Frey JL, Li Z, Ellis JM, Zhang Q, Farber CR, Aja S, et al. Wnt-Lrp5 signaling regulates fatty acid metabolism in the osteoblast. Mol Cell Biol. 2015; 35:1979–1991.
Article123. Frey JL, Kim SP, Li Z, Wolfgang MJ, Riddle RC. β-Catenin directs long-chain fatty acid catabolism in the osteoblasts of male mice. Endocrinology. 2018; 159:272–284.
Article124. Andrade AC, Nilsson O, Barnes KM, Baron J. Wnt gene expression in the post-natal growth plate: regulation with chondrocyte differentiation. Bone. 2007; 40:1361–1369.
Article125. Ayturk UM, Jacobsen CM, Christodoulou DC, Gorham J, Seidman JG, Seidman CE, et al. An RNA-seq protocol to identify mRNA expression changes in mouse diaphyseal bone: applications in mice with bone property altering Lrp5 mutations. J Bone Miner Res. 2013; 28:2081–2093.126. Tan SH, Senarath-Yapa K, Chung MT, Longaker MT, Wu JY, Nusse R. Wnts produced by Osterix-expressing osteolineage cells regulate their proliferation and differentiation. Proc Natl Acad Sci U S A. 2014; 111:E5262–E5271.
Article127. Campbell SE, Febbraio MA. Effect of ovarian hormones on mitochondrial enzyme activity in the fat oxidation pathway of skeletal muscle. Am J Physiol Endocrinol Metab. 2001; 281:E803–E808.
Article128. Herrero P, Soto PF, Dence CS, Kisrieva-Ware Z, Delano DA, Peterson LR, et al. Impact of hormone replacement on myocardial fatty acid metabolism: potential role of estrogen. J Nucl Cardiol. 2005; 12:574–581.
Article129. Hatta H, Atomi Y, Shinohara S, Yamamoto Y, Yamada S. The effects of ovarian hormones on glucose and fatty acid oxidation during exercise in female ovariectomized rats. Horm Metab Res. 1988; 20:609–611.
Article130. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009; 324:1029–1033.
Article131. Karner CM, Esen E, Chen J, Hsu FF, Turk J, Long F. Wnt protein signaling reduces nuclear acetyl-coa levels to suppress gene expression during osteoblast differentiation. J Biol Chem. 2016; 291:13028–13039.
Article132. Biltz RM, Letteri JM, Pellegrino ED, Palekar A, Pinkus LM. Glutamine metabolism in bone. Miner Electrolyte Metab. 1983; 9:125–131.133. Brown PM, Hutchison JD, Crockett JC. Absence of glutamine supplementation prevents differentiation of murine calvarial osteoblasts to a mineralizing phenotype. Calcif Tissue Int. 2011; 89:472–482.
Article134. Karner CM, Esen E, Okunade AL, Patterson BW, Long F. Increased glutamine catabolism mediates bone anabolism in response to WNT signaling. J Clin Invest. 2015; 125:551–562.
Article135. Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997; 77:731–758.
Article136. Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995; 312(Pt 1):163–167.
Article137. Ning X, He J, Shi X, Yu T, Yang G. Wnt3a regulates mitochondrial biogenesis through p38/CREB pathway. Biochem Biophys Res Commun. 2016; 05. 02. DOI: 10.1016/j.bbrc.2016.05.004. [Epub].
Article138. Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010; 24:1507–1518.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Natural Products Targeting Wnt/β-catenin Signaling Pathway
- Wnt signaling in liver regeneration, disease, and cancer
- RNF43 and ZNRF3 in Wnt Signaling - A Master Regulator at the Membrane
- Regulation of Wnt signaling by protein-protein interaction and post-translational modifications
- Negative feedback regulation of Wnt signaling by Gbetagamma-mediated reduction of Dishevelled