Transl Clin Pharmacol.
2019 Mar;27(1):12-18. 10.12793/tcp.2019.27.1.12.
Introduction to in silico model for proarrhythmic risk assessment under the CiPA initiative
- Affiliations
-
- 1Center for Clinical Pharmacology and Biomedical Research Institute, Chonbuk National University Hospital, Jeonju 54907, Republic of Korea. mgkim@jbnu.ac.kr
- 2Department of Pharmacology, School of Medicine, Chonbuk National University, Jeonju 54907, Republic of Korea.
- KMID: 2442241
- DOI: http://doi.org/10.12793/tcp.2019.27.1.12
Abstract
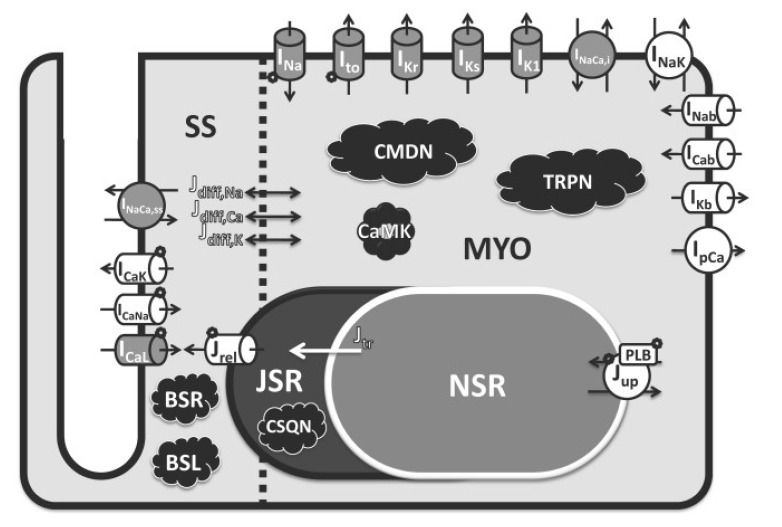
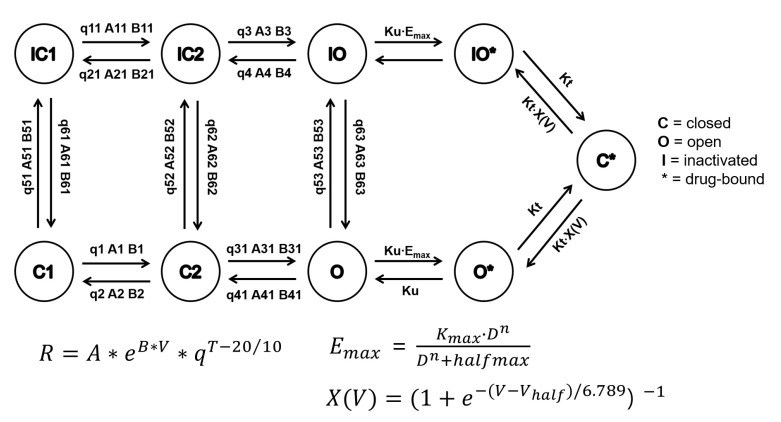
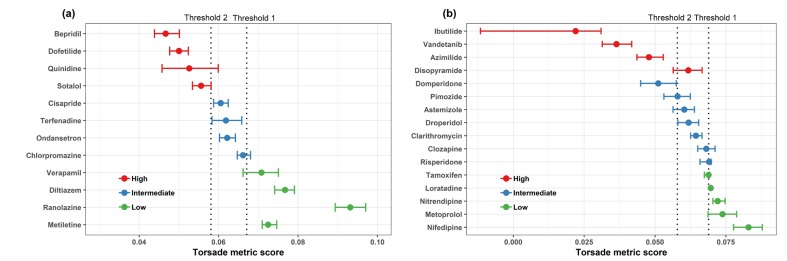
- In 2005, the International Council for Harmonization (ICH) established cardiotoxicity assessment guidelines to identify the risk of Torsade de Pointes (TdP). It is focused on the blockade of the human ether-Ã -go-go-related gene (hERG) channel known to cause QT/QTc prolongation and the QT/QTc prolongation shown on the electrocardiogram. However, these biomarkers are not the direct risks of TdP with low specificity as the action potential is influenced by multiple channels along with the hERG channel. Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative emerged to address limitations of the current model. The objective of CiPA is to develop a standardized in silico model of a human ventricular cell to quantitively evaluate the cardiac response for the cardiac toxicity risk and to come up with a metric for the TdP risk assessment. In silico working group under CiPA developed a standardized and reliable in silico model and a metric that can quantitatively evaluate cellular cardiac electrophysiologic activity. The implementation mainly consists of hERG fitting, Hill fitting, and action potential simulation. In this review, we explained how the in silico model of CiPA works, and briefly summarized current overall CiPA studies. We hope this review helps clinical pharmacologists to understand the underlying estimation process of CiPA in silico modeling.
Keyword
MeSH Terms
Figure
Reference
-
1. Woosley RL, Chen Y, Freiman JP, Gillis RA. Mechanism of the cardiotoxic actions of terfenadine. JAMA. 1993; 269:1532–1536. PMID: 8445816.
Article2. Li M, Ramos LG. Drug-Induced QT Prolongation And Torsades de Pointes. P T. 2017; 42:473–477. PMID: 28674475.3. Food and Drug Administration. HHS. International Conference on Harmonisation; guidance on S7B Nonclinical Evaluation of the Potential for Delayed Ventricular Repolarization (QT Interval Prolongation) by Human Pharmaceuticals; availability. Notice. Fed Regist. 2005; 70:61133–61134. PMID: 16237859.4. Shah RR. Drugs, QTc interval prolongation and final ICH E14 guideline : an important milestone with challenges ahead. Drug Saf. 2005; 28:1009–1028. DOI: 10.2165/00002018-200528110-00003. PMID: 16231954.5. Sager PT, Gintant G, Turner JR, Pettit S, Stockbridge N. Rechanneling the cardiac proarrhythmia safety paradigm: a meeting report from the Cardiac Safety Research Consortium. Am Heart J. 2014; 167:292–300. DOI: 10.1016/j.ahj.2013.11.004. PMID: 24576511.
Article6. Colatsky T, Fermini B, Gintant G, Pierson JB, Sager P, Sekino Y, et al. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative - Update on progress. J Pharmacol Toxicol Methods. 2016; 81:15–20. DOI: 10.1016/j.vascn.2016.06.002. PMID: 27282641.
Article7. Vicente J, Zusterzeel R, Johannesen L, Mason J, Sager P, Patel V, et al. Mechanistic Model-Informed Proarrhythmic Risk Assessment of Drugs: Review of the "CiPA" Initiative and Design of a Prospective Clinical Validation Study. Clin Pharmacol Ther. 2018; 103:54–66. DOI: 10.1002/cpt.896. PMID: 28986934.
Article8. Servick K. A painstaking overhaul for cardiac safety testing. Science. 2016; 353:976–977. PMID: 27701095.
Article9. Fermini B, Hancox JC, Abi-Gerges N, Bridgland-Taylor M, Chaudhary KW, Colatsky T, et al. A New Perspective in the Field of Cardiac Safety Testing through the Comprehensive In Vitro Proarrhythmia Assay Paradigm. J Biomol Screen. 2016; 21:1–11. DOI: 10.1177/1087057115594589. PMID: 26170255.
Article10. About CiPA. Accessed 30 September 2018. http://cipaproject.org/about-cipa/.11. Lancaster MC, Sobie EA. Improved Prediction of Drug-Induced Torsades de Pointes Through Simulations of Dynamics and Machine Learning Algorithms. Clin Pharmacol Ther. 2016; 100:371–379. DOI: 10.1002/cpt.367. PMID: 26950176.
Article12. Mirams GR, Cui Y, Sher A, Fink M, Cooper J, Heath BM, et al. Simulation of multiple ion channel block provides improved early prediction of compounds' clinical torsadogenic risk. Cardiovasc Res. 2011; 91:53–61. DOI: 10.1093/cvr/cvr044. PMID: 21300721.
Article13. O'Hara T, Virag L, Varró A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol. 2011; 7:e1002061. DOI: 10.1371/journal.pcbi.1002061. PMID: 21637795.14. github.com/FDA/CiPA. Accessed 30 September 2018. https://github.com/FDA/CiPA.15. Chang KC, Dutta S, Mirams GR, Beattie KA, Sheng J, Tran PN, et al. Uncertainty Quantification Reveals the Importance of Data Variability and Experimental Design Considerations for in Silico Proarrhythmia Risk Assessment. Front Physiol. 2017; 8:917. DOI: 10.3389/fphys.2017.00917. PMID: 29209226.
Article16. Pathmanathan P, Shotwell MS, Gavaghan DJ, Cordeiro JM, Gray RA. Uncertainty quantification of fast sodium current steady-state inactivation for multi-scale models of cardiac electrophysiology. Prog Biophys Mol Biol. 2015; 117:4–18. DOI: 10.1016/j.pbiomolbio.2015.01.008. PMID: 25661325.
Article17. Li Z, Dutta S, Sheng J, Tran PN, Wu W, Chang K, et al. Improving the In Silico Assessment of Proarrhythmia Risk by Combining hERG (Human Ether-a-go-go-Related Gene) Channel-Drug Binding Kinetics and Multichannel Pharmacology. Circ Arrhythm Electrophysiol. 2017; 10:e004628. DOI: 10.1161/circep.116.004628. PMID: 28202629.
Article18. Dutta S, Chang KC, Beattie KA, Sheng J, Tran PN, Wu WW, et al. Optimization of an In silico Cardiac Cell Model for Proarrhythmia Risk Assessment. Front Physiol. 2017; 8:616. DOI: 10.3389/fphys.2017.00616. PMID: 28878692.
Article19. In Silico Proarrhythmia Risk Assessment under the CiPA Initiative. Accessed 30 September 2018. http://cipaproject.org/wp-content/uploads/sites/24/2017/11/FINALSPS2017AnnualMeeting_CiPAModeling.pdf.20. Li Z, Ridder BJ, Han X, Wu WW, Sheng J, Tran PN, et al. Assessment of an In Silico Mechanistic Model for Proarrhythmia Risk Prediction Under the CiPA Initiative. Clin Pharmacol Ther. 2019; 105:466–475. DOI: 10.1002/cpt.1184. PMID: 30151907.21. Blinova K, Dang Q, Millard D, Smith G, Pierson J, Guo L, et al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell Rep. 2018; 24:3582–3592. DOI: 10.1016/j.celrep.2018.08.079. PMID: 30257217.
Article22. Vicente J, Zusterzeel R, Johannesen L, Ochoa-Jimenez R, Mason JW, Sanabria C, et al. Assessment of Multi-Ion Channel Block in a Phase-1 Randomized Study Design: Results of the CiPA Phase 1 ECG Biomarker Validation Study. Clin Pharmacol Ther. 2018; DOI: 10.1002/cpt.1303.23. Final Concept Paper - ICH S7B and E14 Q&A. Accessed 10 March 2019. https://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Efficacy/E14/E14S7BIWG_ConceptPaper_Final_2018_1122.pdf.24. Strauss DG, Gintant G, Li Z, Wu W, Blinova K, Vicente J, et al. Comprehensive In Vitro Proarrhythmia Assay (CiPA) Update from a Cardiac Safety Research Consortium / Health and Environmental Sciences Institute / FDA Meeting. Ther Innov Regul Sci. 2018; 2168479018795117. DOI: 10.1177/2168479018795117.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Five years of the CiPA project (2013–2018): what did we learn?
- Clinical and pharmacological application of multiscale multiphysics heart simulator, UT-Heart
- A Case of Generalized Xerotic Eczema in a Patient with Congenital Insensitivity to Pain with Anhidrosis
- Anesthetic Management in a Patient with Congenital Insensitivity to Pain with Anhidrosis: A case report
- Anesthetic Management for a Patient with Congenital Insensitivity to Pain with Anhidrosis (CIPA): A case report