J Breast Cancer.
2019 Mar;22(1):15-28. 10.4048/jbc.2019.22.e11.
Pokemon Inhibits Transforming Growth Factor β-Smad4-Related Cell Proliferation Arrest in Breast Cancer through Specificity Protein 1
- Affiliations
-
- 1Institute of Clinical Medicine, the First Affiliated Hospital of University of South China, Hengyang, China. zuxuyu0108@hotmail.com
- 2Division of Stem Cell Regulation and Application, Key Laboratory for Quality Evaluation of Bulk Herbs of Hunan Province, Hunan University of Chinese Medicine, Changsha, China.
- 3Key Laboratory of Carcinogenesis of Ministry of Health and Key Laboratory of Carcinogenesis and Cancer Invasion of Ministry of Education, Cancer Research Institute, Central South University, Changsha, China.
- KMID: 2441848
- DOI: http://doi.org/10.4048/jbc.2019.22.e11
Abstract
- PURPOSE
Pokemon, also known as ZBTB7A, belongs to the POZ and Krüppel (POK) family of transcription repressors and is implicated in tumor progression as a key proto-oncogene. This present study aimed at determining the mechanism by which Pokemon inhibits transforming growth factor β (TGFβ)-Smad4 pathway-dependent proliferation arrest of breast cancer cells via specificity protein 1 (SP1).
METHODS
Over-expressing plasmid or small interfering RNA (siRNA) transfection was used to regulate Pokemon levels. The EdU incorporation assay, MTS assay, and clone formation were used to identify the inhibitory effect of Pokemon siRNA on cell proliferation. Quantitative real-time polymerase chain reaction assay confirmed that Pokemon deletion inhibited the expression of proliferation-associated genes. The dual-luciferase reporter assay, electrophoretic mobility shift assay, and co-immunoprecipitation assay were used to analyze binding between Pokemon, Smad4, and SP1.
RESULTS
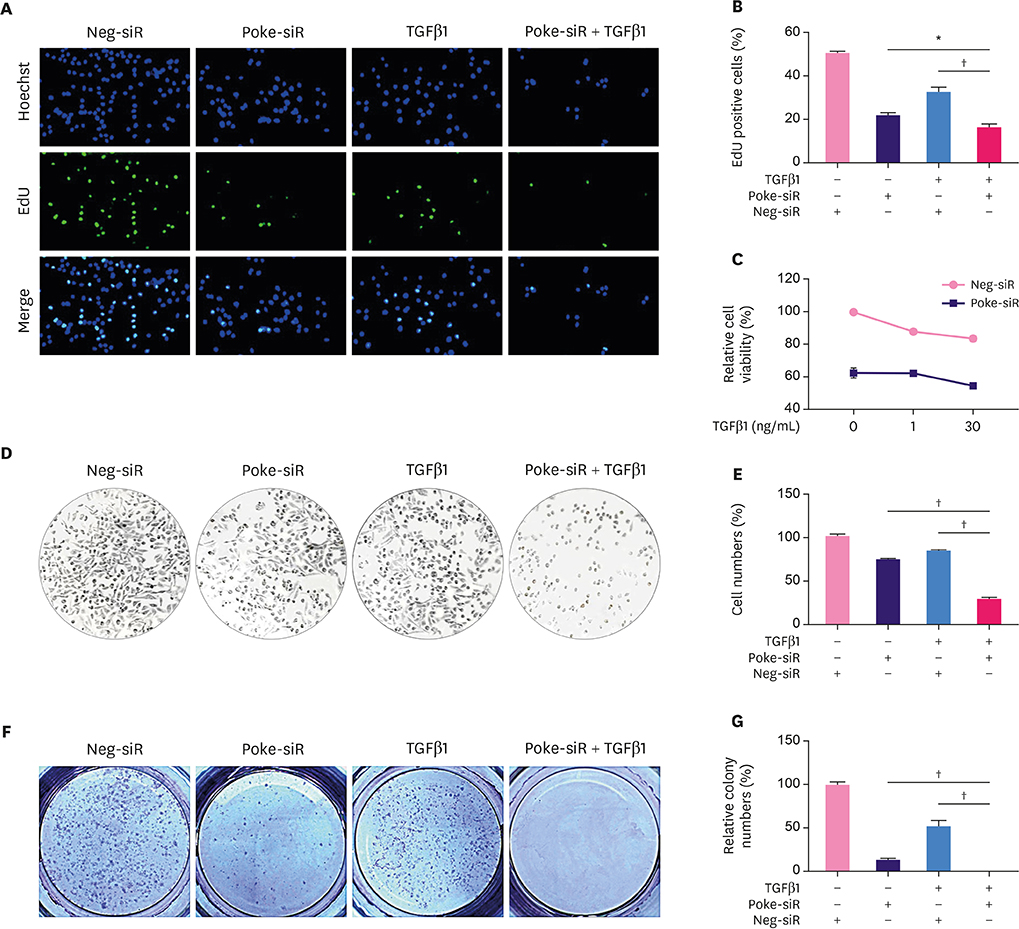
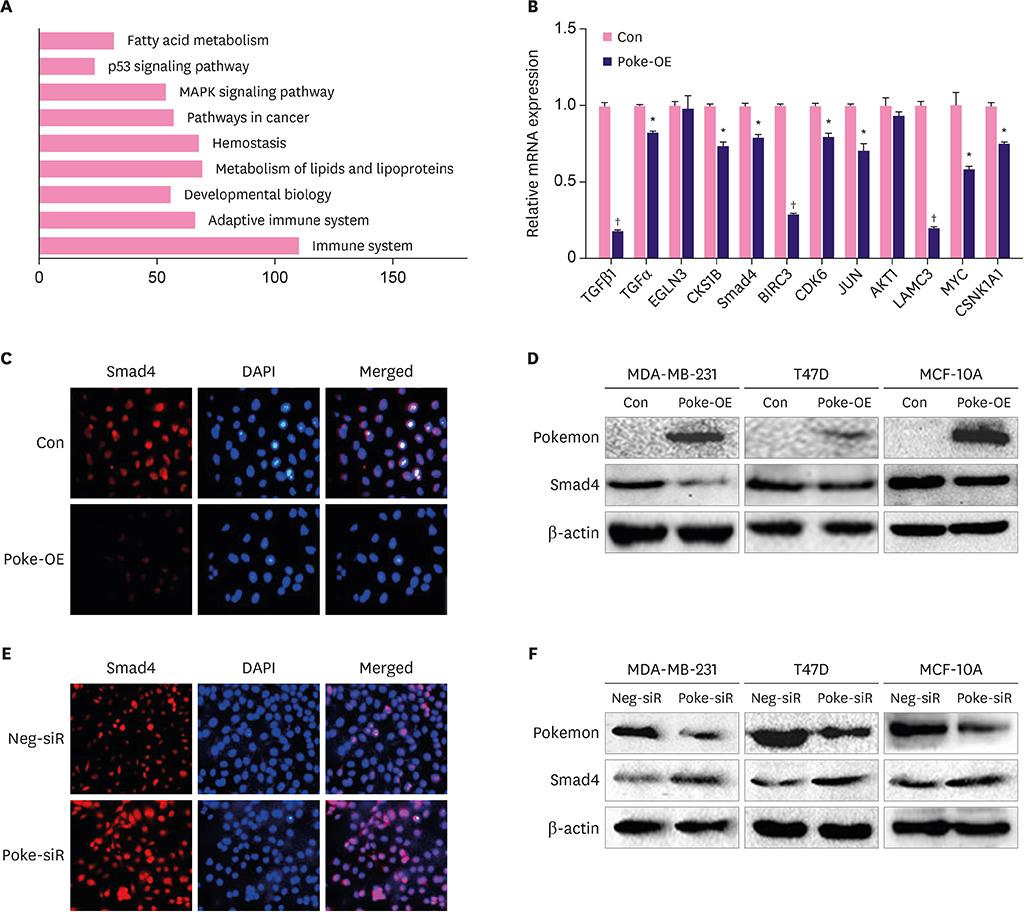
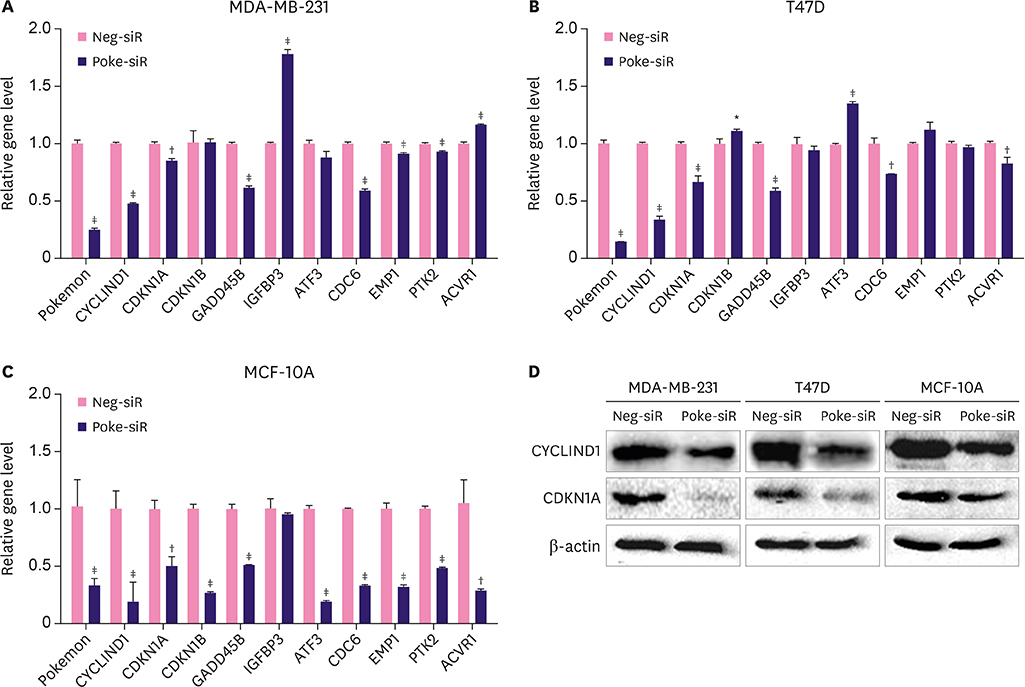
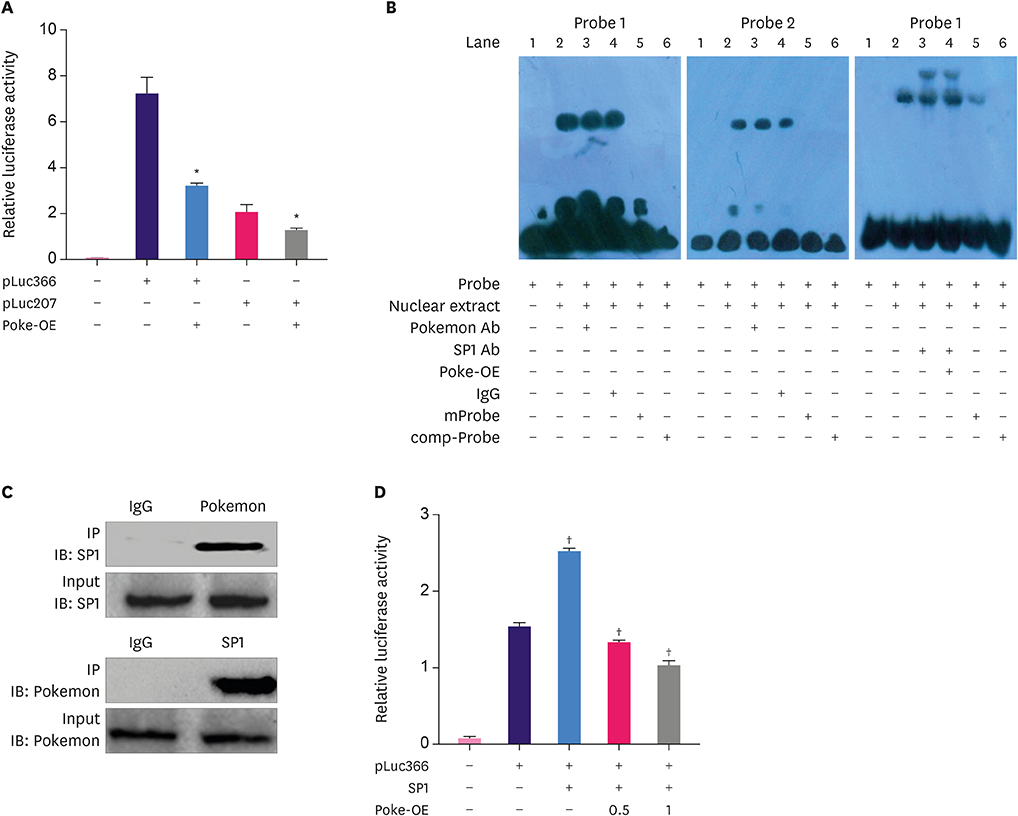
Pokemon deletion induced proliferation arrest of breast cancer cells and inhibited the expression of proliferation-associated genes, especially Smad4. Pokemon bound with SP1 to interdict Smad4 promoter activity. Information on clinical samples was obtained from The Cancer Genome Atlas data, in which the Pokemon mRNA levels showed a negative correlation with Smad4 levels in different subtypes of breast cancer in two independent datasets.
CONCLUSION
We demonstrated that Pokemon binds to SP1 to down-regulate Smad4 expression, thereby promoting proliferation of breast cancer cells. This suggests that Pokemon is a potential TGFβ-signaling participant in breast cancer progression.
MeSH Terms
-
Breast Neoplasms*
Breast*
Cell Proliferation*
Clone Cells
Dataset
Electrophoretic Mobility Shift Assay
Genome
Humans
Immunoprecipitation
Plasmids
Proto-Oncogenes
Real-Time Polymerase Chain Reaction
RNA, Messenger
RNA, Small Interfering
Sensitivity and Specificity*
Smad4 Protein
Transfection
Transforming Growth Factors*
RNA, Messenger
RNA, Small Interfering
Smad4 Protein
Transforming Growth Factors
Figure
Reference
-
1. Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004; 6:17–32.
Article2. Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008; 216:141–150.
Article3. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000; 406:747–752.
Article4. Bae SN, Arand G, Azzam H, Pavasant P, Torri J, Frandsen TL, et al. Molecular and cellular analysis of basement membrane invasion by human breast cancer cells in Matrigel-based in vitro assays. Breast Cancer Res Treat. 1993; 24:241–255.
Article5. Hoogstraat M, Lips E, Mulder L, Nederlof P, Sonke G, Rodenhuis S, et al. Comprehensive characterization of matched pre-treatment biopsies and residual disease of chemotherapy treated breast cancer. Eur J Cancer. 2016; 61:Suppl 1. S30.
Article6. Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006; 10:515–527.
Article7. Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017; 389:2430–2442.
Article8. Moulder SL, Litton JK, Mittendorf E, Yang W, Ueno N, Hess KR, et al. Improving outcomes in triple-negative breast cancer (TNBC) using molecular characterization and diagnostic imaging to identify and treat chemo-insensitive disease. Ann Oncol. 2016; 27:220Tip.
Article9. Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008; 14:1368–1376.
Article10. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363:1938–1948.
Article11. Jeon BN, Yoo JY, Choi WI, Lee CE, Yoon HG, Hur MW. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J Biol Chem. 2008; 283:33199–33210.
Article12. Apostolopoulou K, Pateras IS, Evangelou K, Tsantoulis PK, Liontos M, Kittas C, et al. Gene amplification is a relatively frequent event leading to ZBTB7A (Pokemon) overexpression in non-small cell lung cancer. J Pathol. 2007; 213:294–302.
Article13. Aggarwal A, Hunter WJ 3rd, Aggarwal H, Silva ED, Davey MS, Murphy RF, et al. Expression of leukemia/lymphoma-related factor (LRF/POKEMON) in human breast carcinoma and other cancers. Exp Mol Pathol. 2010; 89:140–148.
Article14. Zu X, Ma J, Liu H, Liu F, Tan C, Yu L, et al. Pro-oncogene Pokemon promotes breast cancer progression by upregulating survivin expression. Breast Cancer Res. 2011; 13:R26.
Article15. Jiang L, Siu MK, Wong OG, Tam KF, Lam EW, Ngan HY, et al. Overexpression of proto-oncogene FBI-1 activates membrane type 1-matrix metalloproteinase in association with adverse outcome in ovarian cancers. Mol Cancer. 2010; 9:318.
Article16. Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005; 433:278–285.
Article17. Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007; 316:860–866.
Article18. Wang G, Lunardi A, Zhang J, Chen Z, Ala U, Webster KA, et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013; 45:739–746.
Article19. Liu XS, Haines JE, Mehanna EK, Genet MD, Ben-Sahra I, Asara JM, et al. ZBTB7A acts as a tumor suppressor through the transcriptional repression of glycolysis. Genes Dev. 2014; 28:1917–1928.
Article20. Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 2010; 10:415–424.
Article21. Derynck R, Akhurst RJ, Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nat Genet. 2001; 29:117–129.
Article22. Li Q, Wu L, Oelschlager DK, Wan M, Stockard CR, Grizzle WE, et al. Smad4 inhibits tumor growth by inducing apoptosis in estrogen receptor-α-positive breast cancer cells. J Biol Chem. 2005; 280:27022–27028.
Article23. Deckers M, van Dinther M, Buijs J, Que I, Löwik C, van der Pluijm G, et al. The tumor suppressor Smad4 is required for transforming growth factor β-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006; 66:2202–2209.
Article24. Yang Y, Cui J, Xue F, Zhang C, Mei Z, Wang Y, et al. Pokemon (FBI-1) interacts with Smad4 to repress TGF-β-induced transcriptional responses. Biochim Biophys Acta. 2015; 1849:270–281.
Article25. Jonckheere N, Van Der Sluis M, Velghe A, Buisine MP, Sutmuller M, Ducourouble MP, et al. Transcriptional activation of the murine Muc5ac mucin gene in epithelial cancer cells by TGF-beta/Smad4 signalling pathway is potentiated by Sp1. Biochem J. 2004; 377:797–808.
Article26. Zhang W, Ou J, Inagaki Y, Greenwel P, Ramirez F. Synergistic cooperation between Sp1 and Smad3/Smad4 mediates transforming growth factor β1 stimulation of α 2(I)-collagen (COL1A2) transcription. J Biol Chem. 2000; 275:39237–39245.
Article27. Cui J, Yang Y, Zhang C, Hu P, Kan W, Bai X, et al. FBI-1 functions as a novel AR co-repressor in prostate cancer cells. Cell Mol Life Sci. 2011; 68:1091–1103.
Article28. Cheng H, Sun X, Li J, He P, Liu W, Meng X. Knockdown of Uba2 inhibits colorectal cancer cell invasion and migration through downregulation of the Wnt/β-catenin signaling pathway. J Cell Biochem. 2018; 119:6914–6925.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinicopathological Significance of SMAD4 Expression in Breast Cancer
- Hypoxia-Inducible Factor 1α Regulates the Transforming Growth Factor β1/SMAD Family Member 3 Pathway to Promote Breast Cancer Progression
- Smad4 Expression in Gastric Adenocarcinoma
- Polyamines Regulate Growth Factor-Induced Protein Phosphorylation in MCF-7 Human Breast Cancer Cells
- Selective B-RAF V600E Inhibitor PLX4032 Inhibits the Growth of Breast Cancer Cell Lines through Cell Cycle Arrest