Cancer Res Treat.
2019 Jan;51(1):300-312. 10.4143/crt.2018.012.
Investigating the Feasibility of Targeted Next-Generation Sequencing to Guide the Treatment of Head and Neck Squamous Cell Carcinoma
- Affiliations
-
- 1Division of Medical Oncology, Department of Internal Medicine, CHA Bundang Medical Center, Seongnam, Korea.
- 2Department of Hemato-Oncology, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- 3Division of Hematology/Oncology, Department of Internal Medicine, Chung-Ang University Hospital, Chung-Ang University College of Medicine, Seoul, Korea.
- 4Department of Pharmacology, Severance Biomedical Science Institute, Yonsei University of College of Medicine, Yonsei Cancer Research Institute, JE-UK Laboratory of Molecular Cancer Therapeutics, Seoul, Korea.
- 5Division of Hematology and Medical Oncology, International St. Mary's Hospital, Catholic Kwandong University College of Medicine, Incheon, Korea.
- 6Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 7Department of Hemato-Oncology, Keimyung University Dongsan Medical Center, Daegu, Korea.
- 8Department of Internal Medicine, Seoul National University Bundang Hospital, Seongnam, Korea.
- 9Department of Internal Medicine, Uijeongbu St. Mary's Hospital, Uijeongbu, Korea.
- 10Department of Medical Oncology, Gachon University Gil Medical Center, Incheon, Korea.
- 11Divison of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea. nobelg@yuhs.ac
- 12HERINGS, The Institute of Advanced Clinical & Biomedical Research, Seoul, Korea.
- 13Division of Medical Oncology, Department of Internal Medicine, Bucheon St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Bucheon, Korea.
- 14Department of Internal Medicine, Incheon St. Mary's Hospital, Incheon, Korea.
- 15Division of Hemato-Oncology, Department of Internal Medicine, Hallym University Kangdong Sacred Heart Hospital, Hallym University College of Medicine, Seoul, Korea.
- 16Department of Internal Medicine, Konyang University Hospital, Daejeon, Korea.
- 17Division of Medical Oncology, Department of Internal Medicine, Korea University Guro Hospital, Korea University College of Medicine, Seoul, Korea.
- 18Rare Cancers Clinic, Center for Specific Organs Cancer, National Cancer Center, Goyang, Korea.
- 19Department of Internal Medicine, SMG-SNU Boramae Hospital, Seoul, Korea.
- 20Department of Hematology-Oncology, Ajou University School of Medicine, Suwon, Korea.
- 21Department of Internal Medicine, Yeungnam University College of Medicine, Daegu, Korea.
- 22Department of Pharmacology, Institute for Cancer Research, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea.
- 23Department of Pathology, SMG-SNU Boramae Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 24Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea.
- 25Department of Internal Medicine, Chungnam National University Hospital, Daejeon, Korea. hjyun@cnuh.co.kr
- 26Department of Biomedical Systems Informatics and Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, Seoul, Korea. swkim@yuhs.ac
- KMID: 2437621
- DOI: http://doi.org/10.4143/crt.2018.012
Abstract
- PURPOSE
Head and neck squamous cell carcinoma (HNSCC) is a deadly disease in which precision medicine needs to be incorporated. We aimed to implement next-generation sequencing (NGS) in determining actionable targets to guide appropriate molecular targeted therapy in HNSCC patients.
MATERIALS AND METHODS
Ninety-three tumors and matched blood samples underwent targeted sequencing of 244 genes using the Illumina HiSeq 2500 platform with an average depth of coverage of greater than 1,000×. Clinicopathological data from patients were obtained from 17 centers in Korea, and were analyzed in correlation with NGS data.
RESULTS
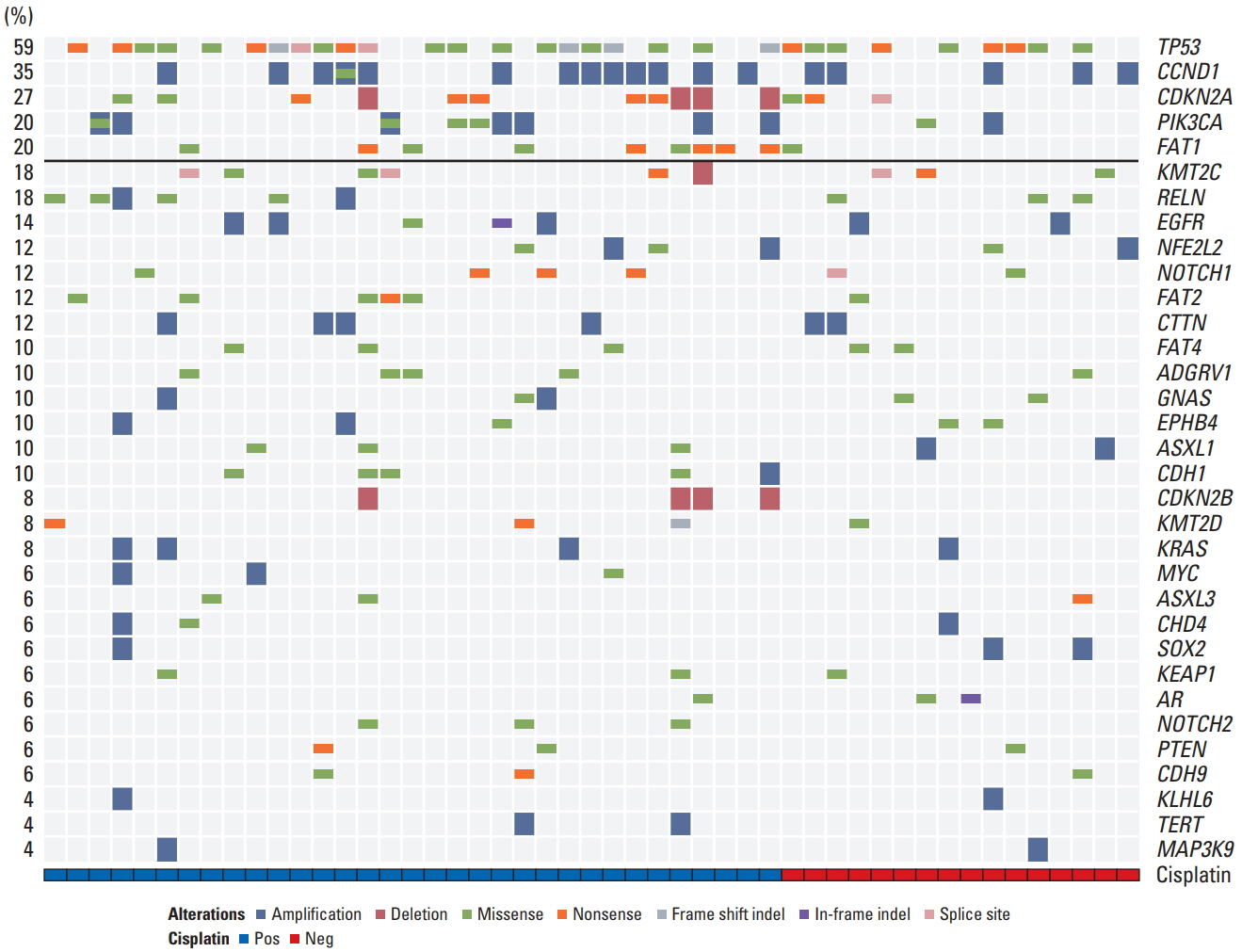
Ninety-two of the 93 tumors were amenable to data analysis. TP53 was the most common mutation, occurring in 47 (51%) patients, followed by CDKN2A (n=23, 25%), CCND1 (n=22, 24%), and PIK3CA (n=19, 21%). The total mutational burden was similar between human papillomavirus (HPV)-negative vs. positive tumors, although TP53, CDKN2A and CCND1 gene alterations occurred more frequently in HPV-negative tumors. HPV-positive tumors were significantly associated with immune signature-related genes compared to HPV-negative tumors. Mutations of NOTCH1 (p=0.027), CDKN2A (p < 0.001), and TP53 (p=0.038) were significantly associated with poorer overall survival. FAT1 mutations were highly enriched in cisplatin responders, and potentially targetable alterations such as PIK3CA E545K and CDKN2A R58X were noted in 14 patients (15%).
CONCLUSION
We found several targetable genetic alterations, and our findings suggest that implementation of precision medicine in HNSCC is feasible. The predictive value of each targetable alteration should be assessed in a future umbrella trial using matched molecular targeted agents.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol. 2007; 25:2171–7.
Article2. Chow LQ, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, et al. Antitumor activity of pembrolizumab in biomarkerunselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 Expansion Cohort. J Clin Oncol. 2016; 34:3838–45.
Article3. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011; 333:1154–7.4. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011; 333:1157–60.
Article5. Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015; 517:576–82.6. Giefing M, Wierzbicka M, Szyfter K, Brenner JC, Braakhuis BJ, Brakenhoff RH, et al. Moving towards personalised therapy in head and neck squamous cell carcinoma through analysis of next generation sequencing data. Eur J Cancer. 2016; 55:147–57.
Article7. Joshi NA, Fass JN. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33) [Internet]. San Francisco, CA: GitHub Inc;2011. [cited 2018 Jan 1]. Available from: https://github.com/najoshi/sickle.8. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–60.
Article9. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–303.
Article10. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013; 31:213–9.
Article11. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012; 22:568–76.
Article12. Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013; 41:e67.
Article13. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.
Article14. Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 2017; 49:170–4.15. Ainscough BJ, Griffith M, Coffman AC, Wagner AH, Kunisaki J, Choudhary MN, et al. DoCM: a database of curated mutations in cancer. Nat Methods. 2016; 13:806–7.
Article16. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genomewide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016; 12:e1004873.
Article17. Waggott DM. NanoString Norm: normalize NanoString miRNA and mRNA data. R Package;2015.18. Wang H, Zhai T, Wang C. NanoStringDiff: differential expression analysis of NanoString nCounter data. R Package;2015.19. Chau NG, Li YY, Jo VY, Rabinowits G, Lorch JH, Tishler RB, et al. Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma. Clin Cancer Res. 2016; 22:2939–49.
Article20. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009; 45:228–47.
Article21. Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol. 2015; 26:1216–23.
Article22. Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015; 21:632–41.
Article23. Massard C, Michiels S, Ferte C, Le Deley MC, Lacroix L, Hollebecque A, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017; 7:586–95.
Article24. Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov. 2015; 14:513–5.
Article25. Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013; 3:761–9.
Article26. Tanoue T, Takeichi M. New insights into Fat cadherins. J Cell Sci. 2005; 118(Pt 11):2347–53.
Article27. Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013; 45:253–61.28. Kim KT, Kim BS, Kim JH. Association between FAT1 mutation and overall survival in patients with human papillomavirusnegative head and neck squamous cell carcinoma. Head Neck. 2016; 38 Suppl 1:E2021–9.29. Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016; 1:e89829.
Article30. Mehra R, Seiwert TY, Gupta S, Weiss J, Gluck I, Eder JP, et al. Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma: pooled analyses after long-term follow-up in KEYNOTE-012. Br J Cancer. 2018; 119:153–9.
Article31. Gillison ML, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab (nivo) vs. investigator's choice (IC) for recurrent or metastatic (R/M) head and neck squamous cell carcinoma (HNSCC): CheckMate-141. In : Proceedings of the 107th Annual Meeting of the American Association for Cancer Research 2016; 2016 Apr 16-20; New Orleans, LA. Philadelphia, PA. American Association for Cancer Research. 2016. Abstr No. CT099.
Article32. Gusnanto A, Wood HM, Pawitan Y, Rabbitts P, Berri S. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics. 2012; 28:40–7.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- EGFR-targeted Therapy in Head and Neck Squamous Cell Carcinoma
- Herpes Viral Gene Therapy for the Treatment of Head and Neck Squamous Cell Carcinoma
- Crosstalk Mechanisms Following Targeted Therapy in Head and Neck Cancer
- Clinical analysis of distant metastases in the squamous cell carcinoma of head and neck
- Expression of p16 Protein and Cyclin D1 Protein in Head and Neck Squamous Cell Carcinomas