Hanyang Med Rev.
2018 Jun;38(2):73-79. 10.7599/hmr.2018.38.2.73.
Human Resistome Study with Metagenomic Sequencing Data
- Affiliations
-
- 1Department of Computer Science and Engineering, College of Engineering, Hanyang University, Seoul, Korea. minarho@hanyang.ac.kr
- 2Department of Biomedical Informatics, Hanyang University, Seoul, Korea.
- KMID: 2431302
- DOI: http://doi.org/10.7599/hmr.2018.38.2.73
Abstract
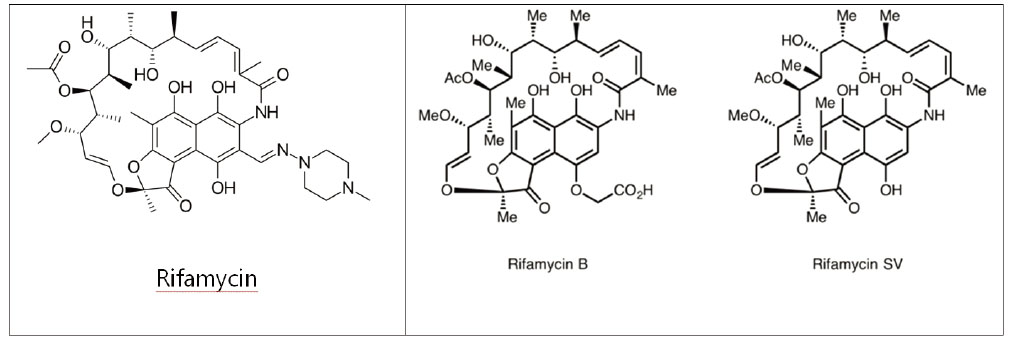
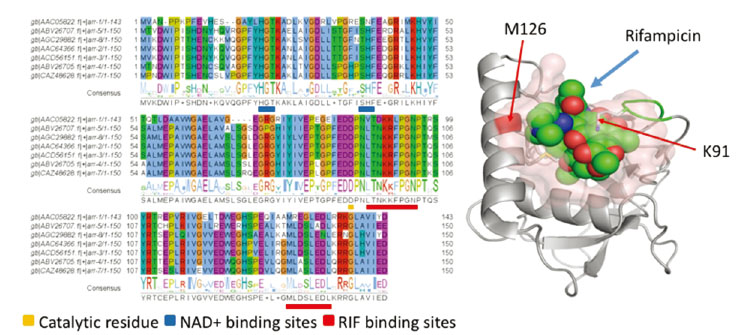
- With the introduction of synthetic antibiotics, many lives including humans and animals have been saved against bacterial infection. An increasing level of antibiotics use, however, raises serious problems of multi-drug resistance and transferring of resistance genes across different environments and countries. Advances in high-throughput sequencing technology and efficient bioinformatics methods allow us to perform a large-scale screening and analysis of resistomes in the human and environmental microbiomes. Recent studies on human microbiomes have revealed a diverse distribution of resistance genes and their transferring activities in the communities. This review discusses recent progresses in metagenomic approaches to identify resistance genes in the human microbiome, including genomic sequence search and functional metagenomics methods. Using Rifampicin ADP-ribosyltransferase as an example, an integrative approach that analyzes the sequences and three-dimensional structures of the proteins derived from resistance genes is also introduced.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Human Microbiome and Resistome Studies
Mina Rho
Hanyang Med Rev. 2018;38(2):71-72. doi: 10.7599/hmr.2018.38.2.71.
Reference
-
1. D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, et al. Antibiotic resistance is ancient. Nature. 2011; 477:457–461.2. Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, et al. Antibiotic Resistance Is Prevalent in an Isolated Cave Microbiome. PLOS ONE. 2012; 7:e34953.
Article3. Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000; 405:299–304.
Article4. Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016; 529:336.
Article5. Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. The Lancet Infectious Diseases. 2014; 14:742–750.
Article6. Spellberg B, Gilbert DN. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis. 2014; 59:Suppl 2. S71–S75.
Article7. Stogios PJ, Cox G, Spanogiannopoulos P, Pillon MC, Waglechner N, Skarina T, et al. Rifampin phosphotransferase is an unusual antibiotic resistance kinase. Nat Commun. 2016; 7:11343.
Article8. Qi X, Lin W, Ma M, Wang C, He Y, He N, et al. Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc Natl Acad Sci U S A. 2016; 113:3803–3808.
Article9. Baysarowich J, Koteva K, Hughes DW, Ejim L, Griffiths E, Zhang K, et al. Rifamycin antibiotic resistance by ADP-ribosylation: Structure and diversity of Arr. Proc Natl Acad Sci U S A. 2008; 105:4886–4891.
Article10. Koch A, Mizrahi V, Warner DF. The impact of drug resistance on Mycobacterium tuberculosis physiology: what can we learn from rifampicin. Emerg Microbes Infect. 2014; 3:e17.11. Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Science Progress. 2002; 85:57–72.
Article12. Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology. 2014; 13:42.
Article13. Knapp CW, Dolfing J, Ehlert PAI, Graham DW. Evidence of Increasing Antibiotic Resistance Gene Abundances in Archived Soils since 1940. Environmental Science & Technology. 2010; 44:580–587.
Article14. Andersson DI, Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev. 2011; 35:901–911.
Article15. Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol. 2001; 67:561–568.
Article16. von Wintersdorff CJH, Penders J, Stobberingh EE, Lashof AMLO, Hoebe CJPA, Savelkoul PHM, et al. High Rates of Antimicrobial Drug Resistance Gene Acquisition after International Travel, the Netherlands. Emerging Infectious Diseases. 2014; 20:649–657.
Article17. Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000; 406:775–781.
Article18. Alekshun MN, Levy SB. Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell. 2007; 128:1037–1050.
Article19. Ventola CL. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharmacy and Therapeutics. 2015; 40:277–283.20. Ventola CL. The Antibiotic Resistance Crisis: Part 2: Management Strategies and New Agents. Pharmacy and Therapeutics. 2015; 40:344–352.21. Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agerso Y, Lund O, et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother. 2013; 68:771–777.
Article22. Anjum MF. Screening methods for the detection of antimicrobial resistance genes present in bacterial isolates and the microbiota. Future Microbiology. 2015; 10:317–320.
Article23. Zhang XX, Zhang T, Fang HH. Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol. 2009; 82:397–414.
Article24. Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003; 57:369–394.
Article25. Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016; 7:13219.
Article26. Adu-Oppong B, Gasparrini AJ, Dantas G. Genomic and functional techniques to mine the microbiome for novel antimicrobials and antimicrobial resistance genes. Ann N Y Acad Sci. 2017; 1388:42–58.
Article27. Antonopoulos DA, Assaf R, Aziz RK, Brettin T, Bun C, Conrad N, et al. PATRIC as a unique resource for studying antimicrobial resistance. Briefings in Bioinformatics. 2017; bbx083-bbx.
Article28. Lee D, Das S, Dawson NL, Dobrijevic D, Ward J, Orengo C. Novel Computational Protocols for Functionally Classifying and Characterising Serine Beta-Lactamases. PLOS Computational Biology. 2016; 12:e1004926.
Article29. Philippon A, Slama P, Dény P, Labia R. A Structure-Based Classification of Class A β-Lactamases, a Broadly Diverse Family of Enzymes. Clinical Microbiology Reviews. 2016; 29:29–57.
Article30. Lam KN, Cheng J, Engel K, Neufeld JD, Charles TC. Current and future resources for functional metagenomics. Front Microbiol. 2015; 6:1196.
Article31. Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017; 15:422–434.
Article32. dos Santos DFK, Istvan P, Quirino BF, Kruger RH. Functional Metagenomics as a Tool for Identification of New Antibiotic Resistance Genes from Natural Environments. Microbial Ecology. 2017; 73:479–491.
Article33. Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009; 325:1128–1131.
Article34. Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004; 6:981–989.
Article35. Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. The ISME journal. 2009; 3:243–251.
Article36. Liu B, Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009; 37:D443–D447.
Article37. McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013; 57:3348–3357.
Article38. Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. Isme j. 2015; 9:207–216.
Article39. Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017; 45:D566–Dd73.
Article40. Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. The ISME Journal. 2015; 9:207–216.
Article41. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012; 67:2640–2644.
Article42. Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014; 58:212–220.
Article43. Arango-Argoty G, Garner E, Pruden A, Heath LS, Vikesland P, Zhang L. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018; 6:23.
Article44. Hartmann G, Honikel KO, Knüsel F, Nüesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967; 145:843–844.
Article45. Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016; 4:DOI: 10.1128/microbiolspec.VMBF-0016-2015.
Article46. Stogios PJ, Cox G, Spanogiannopoulos P, Pillon MC, Waglechner N, Skarina T, et al. Rifampin phosphotransferase is an unusual antibiotic resistance kinase. Nat Commun. 2016; 7:11343.
Article47. Baysarowich J, Koteva K, Hughes DW, Ejim L, Griffiths E, Zhang K, et al. Rifamycin antibiotic resistance by ADP-ribosylation: Structure and diversity of Arr. Proc Natl Acad Sci U S A. 2008; 105:4886–4891.
Article48. Dabbs ER, Yazawa K, Mikami Y, Miyaji M, Morisaki N, Iwasaki S, et al. Ribosylation by mycobacterial strains as a new mechanism of rifampin inactivation. Antimicrob Agents Chemother. 1995; 39:1007–1009.
Article49. Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013; 4:2151.
Article50. Ghosh TS, Gupta SS, Nair GB, Mande SS. In Silico Analysis of Antibiotic Resistance Genes in the Gut Microflora of Individuals from Diverse Geographies and Age-Groups. PLOS ONE. 2013; 8:e83823.
Article51. Yang Z, Guo Z, Qiu C, Li Y, Feng X, Liu Y, et al. Preliminary analysis showed country-specific gut resistome based on 1267 feces samples. Gene. 2016; 581:178–182.
Article52. Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, et al. The Human Gut Microbiome as a Transporter of Antibiotic Resistance Genes between Continents. Antimicrobial Agents and Chemotherapy. 2015; 59:6551.
Article53. Gibson MK, Wang B, Ahmadi S, Burnham C-AD, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016; 1:16024.
Article54. Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, et al. Differential Effects of Antibiotic Therapy on the Structure and Function of Human Gut Microbiota. PLOS ONE. 2013; 8:e80201.
Article55. Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Barnabas B, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014; 514:59.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Application of Metagenomic Techniques: Understanding the Unrevealed Human Microbiota and Explaining the in Clinical Infectious Diseases
- Analytical Tools and Databases for Metagenomics in the Next-Generation Sequencing Era
- Human Microbiome and Resistome Studies
- Changes in serum circulating bacterial DNA fragment before and after kidney transplantation and its clinical significance
- Subgingival microbiome in periodontitis and type 2 diabetes mellitus: an exploratory study using metagenomic sequencing