Yonsei Med J.
2019 Jan;60(1):22-29. 10.3349/ymj.2019.60.1.22.
MiR-506 Promotes Natural Killer Cell Cytotoxicity against Human Hepatocellular Carcinoma Cells by Targeting STAT3
- Affiliations
-
- 1Department of General Surgery, Second Affiliated Hospital of Guangxi Medical University, Nanning, China. y2521790543@163.com
- 2Department of Hepatobiliary Surgery, First Affiliated Hospital of Guangxi Medical University, Nanning, China.
- KMID: 2428274
- DOI: http://doi.org/10.3349/ymj.2019.60.1.22
Abstract
- PURPOSE
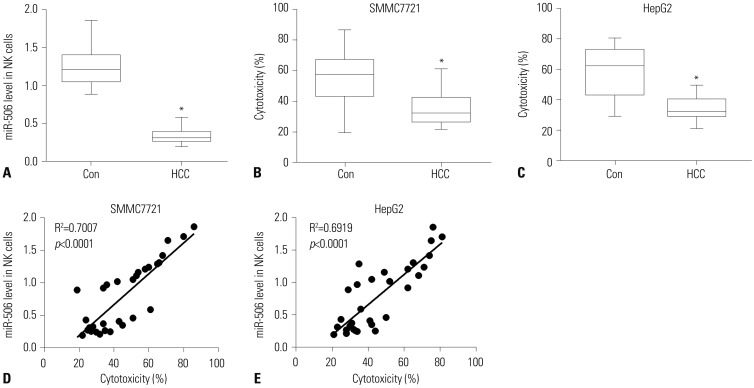
It is well documented that natural killer (NK) cytotoxicity against hepatocellular carcinoma (HCC) cells is impaired in HCC, which might account for the failure of anti-tumor immune response. miRNAs are considered as important regulators for the development and functions of NK cells. However, the entire role of miR-506 in NK cells remains far from being addressed.
MATERIALS AND METHODS
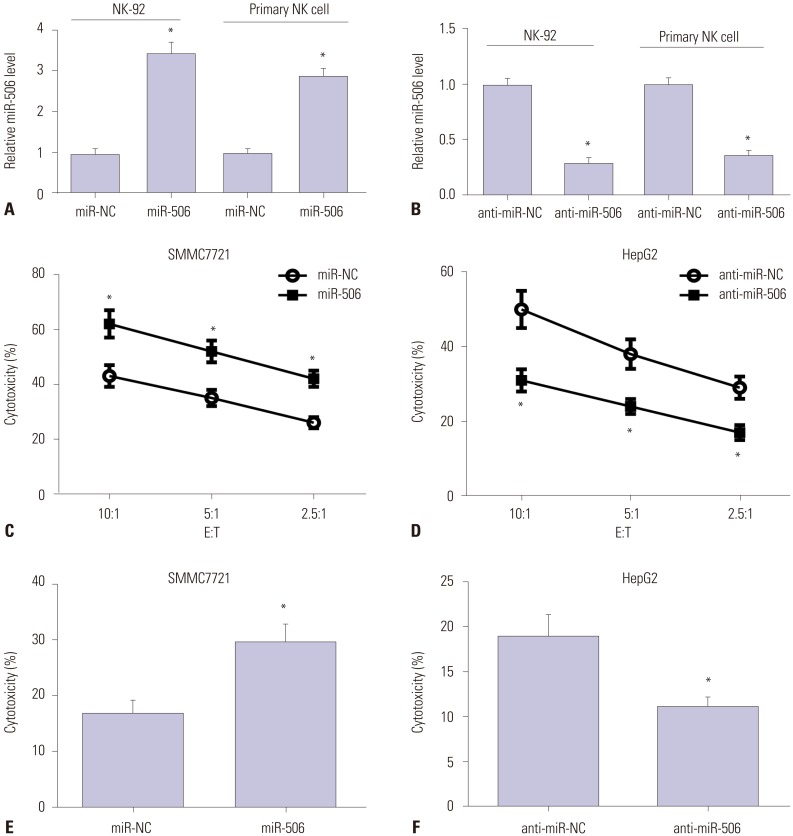
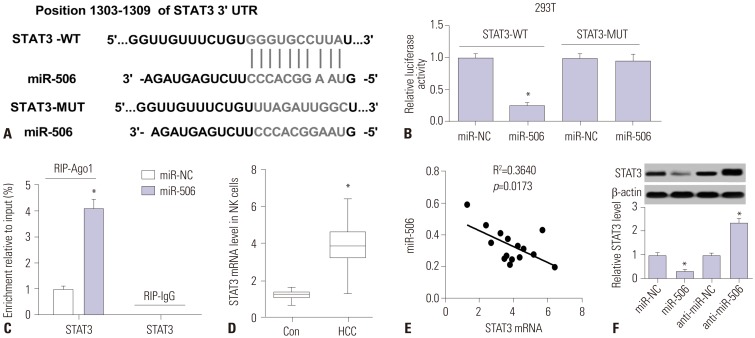
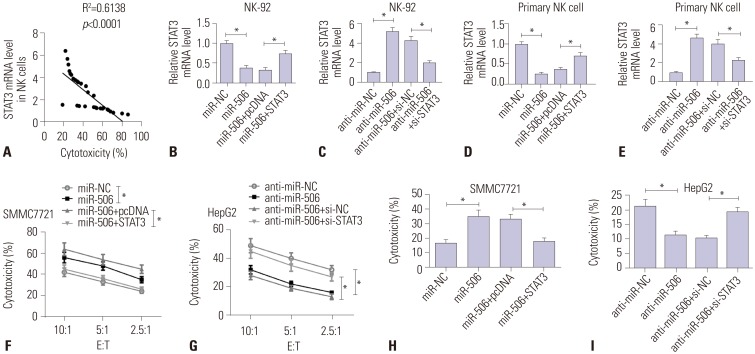
The expressions of miR-506 and signal transducer and activator of transcription 3 (STAT3) mRNA in primary NK cells from HCC patients and healthy controls were detected by quantitative real-time PCR. NK cell cytotoxicity was assessed by CFSE/7AAD cytotoxicity assay and lactate dehydrogenase assay. Luciferase reporter assay, RNA immunoprecipitation assay, and western blot were conducted to confirm the interaction between miR-506 and STAT3.
RESULTS
miR-506 expression was downregulated and STAT3 mRNA was upregulated in primary NK cells from HCC patients. Primary NK cells from HCC patients showed remarkably reduced cytotoxicity against SMMC7721 or HepG2 cells. NK cell cytotoxicity was positively correlated with miR-506 expression and negatively correlated with STAT3 mRNA expression. Additionally, miR-506 overexpression enhanced NK cell cytotoxicity against HCC cells, while miR-506 inhibitor showed the reverse effect. Moreover, miR-506 could suppress STAT3 expression by directly targeting 3"²-untranslated regions of STAT3. A negative correlation between miR-506 and STAT3 mRNA expression in HCC patients was observed. Mechanistically, overexpressing STAT3 greatly reversed miR-506-mediated promotion of NK cell cytotoxicity against HCC cells.
CONCLUSION
miR-506 enhanced NK cell cytotoxicity against HCC cells by targeting STAT3, suggesting that modulating miR-506 expression maybe a promising approach for enhancing NK cell-based antitumor therapies.
Keyword
MeSH Terms
-
Blotting, Western
Carcinoma, Hepatocellular*
Hep G2 Cells
Humans*
Immunoprecipitation
Killer Cells, Natural*
L-Lactate Dehydrogenase
Luciferases
MicroRNAs
Real-Time Polymerase Chain Reaction
RNA
RNA, Messenger
STAT3 Transcription Factor
L-Lactate Dehydrogenase
Luciferases
MicroRNAs
RNA
RNA, Messenger
STAT3 Transcription Factor
Figure
Reference
-
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. PMID: 25651787.
Article2. Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008; 27:5932–5943. PMID: 18836474.
Article3. Ishiyama K, Ohdan H, Ohira M, Mitsuta H, Arihiro K, Asahara T. Difference in cytotoxicity against hepatocellular carcinoma between liver and periphery natural killer cells in humans. Hepatology. 2006; 43:362–372. PMID: 16440347.
Article4. Subleski JJ, Hall VL, Back TC, Ortaldo JR, Wiltrout RH. Enhanced antitumor response by divergent modulation of natural killer and natural killer T cells in the liver. Cancer Res. 2006; 66:11005–11012. PMID: 17108139.
Article5. Zhang QF, Yin WW, Xia Y, Yi YY, He QF, Wang X, et al. Liver-infiltrating CD11b-CD27- NK subsets account for NK-cell dysfunction in patients with hepatocellular carcinoma and are associated with tumor progression. Cell Mol Immunol. 2017; 14:819–829. PMID: 27321064.
Article6. Hoechst B, Voigtlaender T, Ormandy L, Gamrekelashvili J, Zhao F, Wedemeyer H, et al. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology. 2009; 50:799–807. PMID: 19551844.
Article7. Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009; 16:144–150. PMID: 19182800.
Article8. Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016; 76:3666–3670. PMID: 27325641.
Article9. O'Connell RM, Baltimore D. MicroRNAs and hematopoietic cell development. Curr Top Dev Biol. 2012; 99:145–174. PMID: 22365738.10. Squadrito ML, Etzrodt M, De Palma M, Pittet MJ. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013; 34:350–359. PMID: 23498847.
Article11. Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005; 37:766–770. PMID: 15965474.
Article12. Li J, Ju J, Ni B, Wang H. The emerging role of miR-506 in cancer. Oncotarget. 2016; 7:62778–62788. PMID: 27542202.
Article13. Kumar Kingsley SM, Vishnu Bhat B. Role of MicroRNAs in the development and function of innate immune cells. Int Rev Immunol. 2017; 36:154–175. PMID: 28471289.
Article14. Xie H, Zhang Q, Zhou H, Zhou J, Zhang J, Jiang Y, et al. microRNA-889 is downregulated by histone deacetylase inhibitors and confers resistance to natural killer cytotoxicity in hepatocellular carcinoma cells. Cytotechnology. 2018; 70:513–521. PMID: 28550492.
Article15. Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. 2014; 11:495–502. PMID: 24813230.
Article16. Xu D, Han Q, Hou Z, Zhang C, Zhang J. miR-146a negatively regulates NK cell functions via STAT1 signaling. Cell Mol Immunol. 2017; 14:712–720. PMID: 26996068.
Article17. Yu Z, Zhang Y, Gao N, Wang X. Overexpression of miR-506 inhibits growth of osteosarcoma through Snail2. Am J Transl Res. 2015; 7:2716–2723. PMID: 26885269.18. Yao WJ, Wang YL, Lu JG, Guo L, Qi B, Chen ZJ. MicroRNA-506 inhibits esophageal cancer cell proliferation via targeting CREB1. Int J Clin Exp Pathol. 2015; 8:10868–10874. PMID: 26617801.19. Arora H, Qureshi R, Park WY. miR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS One. 2013; 8:e64273. PMID: 23717581.
Article20. Dai W, Huang HL, Hu M, Wang SJ, He HJ, Chen NP, et al. microRNA-506 regulates proliferation, migration and invasion in hepatocellular carcinoma by targeting F-spondin 1 (SPON1). Am J Cancer Res. 2015; 5:2697–2707. PMID: 26609477.21. Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012; 31:1558–1570. PMID: 21860416.
Article22. Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM, Xiao SD, et al. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Lett. 2011; 585:3560–3568. PMID: 22036718.
Article23. Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007; 7:41–51. PMID: 17186030.
Article24. Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004; 10:48–54. PMID: 14702634.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Correlation between NK Cell and Liver Function in Patients with Primary Hepatocellular Carcinoma
- Targeting TM4SF1 to overcome immunotherapy resistance in hepatocellular carcinoma: Editorial on “Targeting TM4SF1 promotes tumor senescence enhancing CD8+ T cell cytotoxic function in hepatocellular carcinoma”
- Establishment of Antibody-dependant Cellular Cytotoxicity against Human Squamous Cell Carcinoma of Head and Neck and The Effect of Prostaglandin on Antibody-dependant Cellular Cytotoxicity
- MiR-218-5p Suppresses the Killing Effect of Natural Killer Cell to Lung Adenocarcinoma by Targeting SHMT1
- Tetraspan(in)-mediated immune regulation in hepatocellular carcinoma: Editorial on “Targeting TM4SF1 promotes tumor senescence enhancing CD8+ T cell cytotoxic function in hepatocellular carcinoma”