Ann Lab Med.
2019 Mar;39(2):167-175. 10.3343/alm.2019.39.2.167.
Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China
- Affiliations
-
- 1Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China. lijiabin948@vip.sohu.com
- 2Department of Laboratory Medicine, The Second Hospital of Anhui Medical University, Hefei, Anhui, China. shiheguan@126.com
- 3Anhui Center for Surveillance of Bacterial Resistance, Hefei, Anhui, China.
- 4Department of Infectious Diseases, Chaohu Hospital of Anhui Medical University, Hefei, Anhui, China.
- KMID: 2425972
- DOI: http://doi.org/10.3343/alm.2019.39.2.167
Abstract
- BACKGROUND
Carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae (CR-HMKP) poses a significant public health challenge. We investigated its epidemiology and molecular characteristics in a tertiary care hospital in eastern China.
METHODS
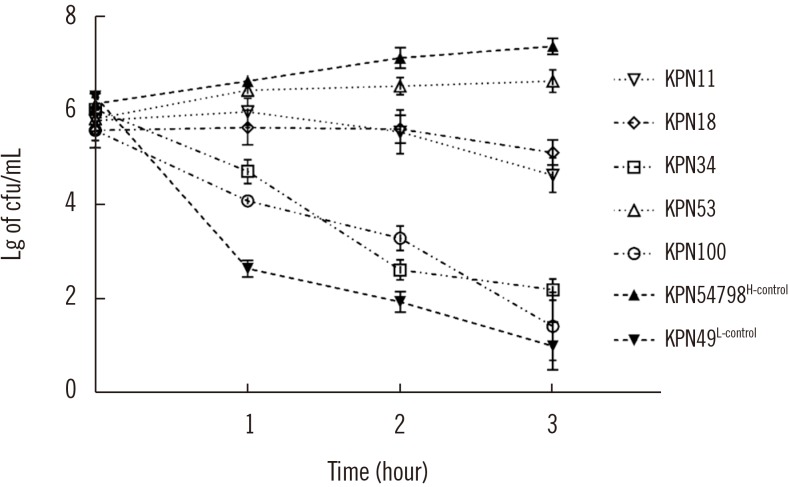
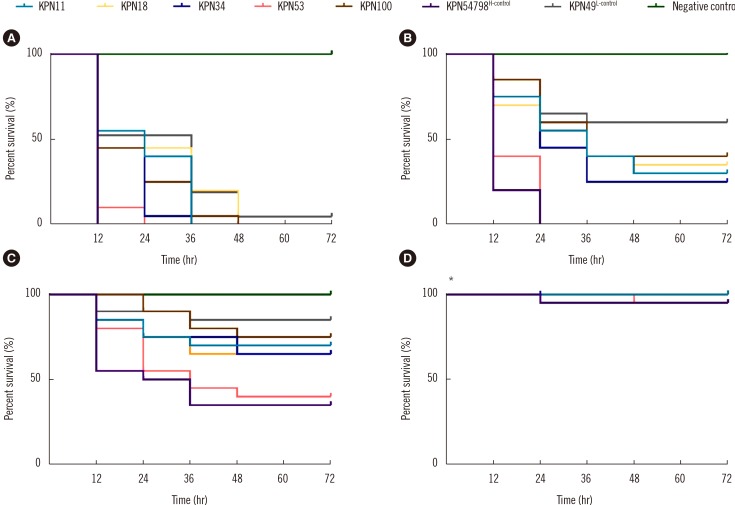
CR-HMKP were identified among 106 non-duplicated carbapenem-resistant K. pneumoniae isolates (from June 2013 to September 2017) using the string test. The pulsotype (PT) and sequence type (ST) of CR-HMKP isolates were determined using pulsed-field gel electrophoresis and multilocus sequence typing. Resistance determinants, capsular serotypes, and virulence genes were detected by PCR and sequencing. Representative isolates from each PT were selected, and their virulence phenotypes were established using the serum killing and Galleria mellonella lethality assays.
RESULTS
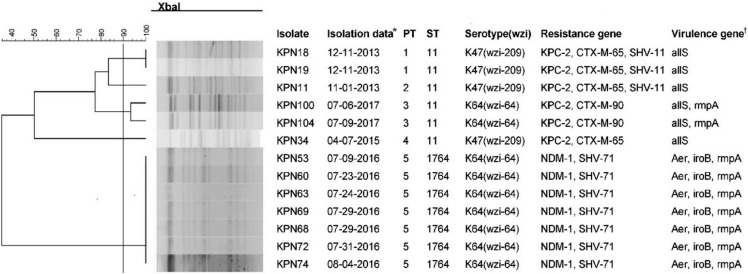
Of the 106 isolates, 13 (12.3%) were CR-HMKP. Seven were positive for bla NDM-1 and shared the same genotype (PT5/ST1764); the others were positive for bla KPC-2, belonged to ST11, and were divided into four different PTs. The serotype of all bla NDM-1-positive isolates was K64, while that of bla KPC-2-positive isolates were K47 (N=4) and K64 (N=2). The NDM-1-producing HMKP isolates were positive for aerobactin, exhibited high serum resistance, and elicited significantly increased larval mortality compared with the other isolates. All patients had received invasive treatment prior to infection by NDM-1-producing HMKP. The infections occurred between July and August 2016 and were hospital-acquired.
CONCLUSIONS
NDM-1-producing HMKP ST1764 isolates were identified; this is the first report worldwide on an outbreak of nosocomial infection caused by these isolates. Effective surveillance and strict infection control strategies should be implemented to prevent CR-HMKP dissemination.
Keyword
MeSH Terms
Figure
Reference
-
1. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017; 41:252–275. PMID: 28521338.2. Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013; 4:107–118. PMID: 23302790.3. Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017; 7:483. PMID: 29209595.4. Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015; 83:3325–3333. PMID: 26056379.5. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016; 80:629–661. PMID: 27307579.6. Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis. 2018; 18:2–3. PMID: 28864028.7. Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017; 7:182. PMID: 28560183.8. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018; 18:37–46. PMID: 28864030.9. Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2015; 60:709–711. PMID: 26574010.10. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015; 37:107–112. PMID: 26141415.11. Arena F, Henrici De, D'Andrea MM, Cannatelli A, Fossati L, Di Pilato V, et al. Infections caused by carbapenem-resistant Klebsiella pneumoniae with hypermucoviscous phenotype: a case report and literature review. Virulence. 2017; 8:1900–1908. PMID: 28276993.12. Shankar C, Nabarro LE, Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Daniel JL, Doss C GP, et al. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announc. 2016; 4:pii: e01081-16.13. CLSI. CLSI supplement M100. Performance standards for antimicrobial susceptibility testing. 28th ed. Wayne, PA: Clinical and Laboratory Standards Institute;2018.14. CLSI. CLSI Standard M07. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 11th ed. Wayne, PA: Clinical and Laboratory Standards Institute;2018.15. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.0. 2017. Updated on Jan 2017. http://www.eucast.org.16. Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H. Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother. 2008; 61:548–553. PMID: 18222954.17. Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012; 50:3877–3880. PMID: 22993175.18. Yan J, Pu S, Jia X, Xu X, Yang S, Shi J, et al. Multidrug resistance mechanisms of carbapenem resistant Klebsiella pneumoniae strains isolated in Chongqing, China. Ann Lab Med. 2017; 37:398–407. PMID: 28643488.19. Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013; 51:4073–4078. PMID: 24088853.20. Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, et al. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One. 2013; 8:e80670. PMID: 24349011.21. Liu Y, Liu PP, Wang LH, Wei DD, Wan LG, Zhang W. Capsular polysaccharide types and virulence-related traits of epidemic KPC-producing Klebsiella pneumoniae isolates in a Chinese university Hospital. Microb Drug Resist. 2017; 23:901–907. PMID: 28437231.22. Yan Q, Zhou M, Zou M, Liu WE. Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis. 2016; 35:387–396. PMID: 26753990.23. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43:4178–4182. PMID: 16081970.24. Chiang TT, Yang YS, Yeh KM, Chiu SK, Wang NC, Lin TY, et al. Quantification and comparison of virulence and characteristics of different variants of carbapenemase-producing Klebsiella pneumoniae clinical isolates from Taiwan and the United States. J Microbiol Immunol Infect. 2016; 49:83–90. PMID: 26514941.25. Mclaughlin MM, Advincula MR, Malczynski M, Barajas G, Qi C, Scheetz MH. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a Galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect Dis. 2014; 14:31. PMID: 24428847.26. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008; 36:309–332. PMID: 18538699.27. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016; 7:895. PMID: 27379038.28. Mei YF, Liu PP, Wan LG, Liu Y, Wang LH, Wei DD, et al. Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence Type 14 strain of serotype K2 harboring blaNDM-5 in China. Front Microbiol. 2017; 8:335. PMID: 28386246.29. Compain F, Vandenberghe A, Gominet M, Genel N, Lebeaux D, Ramahefasolo A, et al. Primary osteomyelitis caused by an NDM-1-producing K. pneumoniae strain of the highly virulent sequence type 23. Emerg Microbes Infect. 2017; 6:e57. PMID: 28634354.30. Wei DD, Wan LG, Liu Y. Draft genome sequence of an NDM-1- and KPC-2-coproducing hypervirulent carbapenem-resistant Klebsiella pneumoniae strain isolated from burn wound infections. Genome Announc. 2018; 6:pii: e00192-18.31. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011; 66:307–312. PMID: 21131324.32. Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL. The function of wzy_K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J Infect Dis. 2010; 201:1268–1269. PMID: 20225957.33. Liu YM, Li BB, Zhang YY, Zhang W, Shen H, Li H, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014; 58:5379–5385. PMID: 24982067.34. Sun Y, Wu H, Shen D. Clinical and molecular analysis of Klebsiella pneumoniae causing liver abscess in China. J Mol Microbiol Biotechnol. 2016; 26:245–251. PMID: 27073997.35. Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014; 82:2356–2367. PMID: 24664504.36. Wand ME, McCowen JW, Nugent PG, Sutton JM. Complex interactions of Klebsiella pneumoniae with the host immune system in a Galleria mellonella infection model. J Med Microbiol. 2013; 62:1790–1798. PMID: 24000226.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Central Nervous System Infection with Hypervirulent Klebsiella pneumoniae
- Rapid Identification of OXA-48-like, KPC, NDM, and VIM Carbapenemase-Producing Enterobacteriaceae From Culture: Evaluation of the RESIST-4 O.K.N.V. Multiplex Lateral Flow Assay
- The First Klebsiella pneumoniae Isolate Co-Producing OXA-48 and NDM-1 in Turkey
- Necrotizing fasciitis and psoas abscess caused by hypervirulent Klebsiella pneumoniae
- In Vitro Susceptibility of piperacillin/tazobactam Against extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae