Obstet Gynecol Sci.
2018 Mar;61(2):235-241. 10.5468/ogs.2018.61.2.235.
Sequencing analysis of HPV-other type on an HPV DNA chip
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Healthcare Research Institute, Gangnam Healthcare Center, Seoul National University Hospital, Seoul, Korea. sunmie@snuh.org
- KMID: 2420177
- DOI: http://doi.org/10.5468/ogs.2018.61.2.235
Abstract
OBJECTIVES
To identify the specific human papillomavirus (HPV) genotypes from HPV-other type on an HPV DNA chip test by sequencing.
METHODS
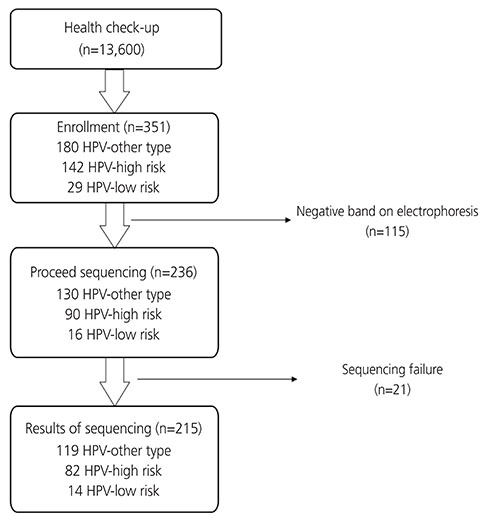
Among 13,600 women undergoing a routine gynecology examination including Pap smear and/or HPV test by DNA chip test in the healthcare system at Gangnam Center from July 2012 to February 2013, we prospectively collected and performed sequencing for a total of 351 consecutive cervicovaginal samples consisting of 180 samples that tested positive for HPV-other type and 171 samples that tested positive for either high-risk HPV or low-risk HPV.
RESULTS
Of a total of 351 samples, individual HPV genotypes were successfully sequenced in 215 cases: 119 HPV-other type, 82 HPV-high-risk, and 14 HPV-low-risk. Based on the sequencing for 119 HPV-other type samples, 91.6% were detected as HPV types that were not included on the DNA chip; however, 7.6% (9/119) were proven to be high-risk HPV types: HPV 18 (n=4), HPV 33 (n=3), HPV 35 (n=1), and HPV 59 (n=1). For correlation analysis of all high-risk and HPV 16/18, the correlation rate was 76.2% and 86.6% with kappa-value of 0.38 and 0.69, respectively.
CONCLUSION
HPV-other type on DNA chip test may still have possibility of high-risk HPV, i.e., HPV 18 and thus the significance of HPV-other type in detecting cervical disease remains to be investigated.
MeSH Terms
Figure
Reference
-
1. Clifford GM, Rana RK, Franceschi S, Smith JS, Gough G, Pimenta JM. Human papillomavirus genotype distribution in low-grade cervical lesions: comparison by geographic region and with cervical cancer. Cancer Epidemiol Biomarkers Prev. 2005; 14:1157–1164.
Article2. Naucler P, Ryd W, Törnberg S, Strand A, Wadell G, Hansson BG, et al. HPV type-specific risks of high-grade CIN during 4 years of follow-up: a population-based prospective study. Br J Cancer. 2007; 97:129–132.
Article3. Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005; 97:1072–1079.
Article4. Bosch FX, de Sanjosé S. Chapter 1: human papillomavirus and cervical cancer--burden and assessment of causality. J Natl Cancer Inst Monogr. 2003; 3–13.
Article5. An HJ, Cho NH, Lee SY, Kim IH, Lee C, Kim SJ, et al. Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with the HPV DNA chip microarray method. Cancer. 2003; 97:1672–1680.
Article6. Oh TJ, Kim CJ, Woo SK, Kim TS, Jeong DJ, Kim MS, et al. Development and clinical evaluation of a highly sensitive DNA microarray for detection and genotyping of human papillomaviruses. J Clin Microbiol. 2004; 42:3272–3280.
Article7. Lee JK, Kim MK, Song SH, Hong JH, Min KJ, Kim JH, et al. Comparison of human papillomavirus detection and typing by hybrid capture 2, linear array, DNA chip, and cycle sequencing in cervical swab samples. Int J Gynecol Cancer. 2009; 19:266–272.
Article8. Hwang TS, Jeong JK, Park M, Han HS, Choi HK, Park TS. Detection and typing of HPV genotypes in various cervical lesions by HPV oligonucleotide microarray. Gynecol Oncol. 2003; 90:51–56.
Article9. Choi YD, Jung WW, Nam JH, Choi HS, Park CS. Detection of HPV genotypes in cervical lesions by the HPV DNA Chip and sequencing. Gynecol Oncol. 2005; 98:369–375.
Article10. Um TH, Lee EH, Chi HS, Kim JW, Hong YJ, Cha YJ. Comparison of HPV genotyping assays and Hybrid Capture 2 for detection of high-risk HPV in cervical specimens. Ann Clin Lab Sci. 2011; 41:48–55.11. Park S, Kang Y, Kim DG, Kim EC, Park SS, Seong MW. Comparison of the analytical and clinical performances of Abbott RealTime High Risk HPV, Hybrid Capture 2, and DNA Chip assays in gynecology patients. Diagn Microbiol Infect Dis. 2013; 76:432–436.
Article12. Klug SJ, Molijn A, Schopp B, Holz B, Iftner A, Quint W, et al. Comparison of the performance of different HPV genotyping methods for detecting genital HPV types. J Med Virol. 2008; 80:1264–1274.
Article13. Kim MJ, Kim JJ, Kim S. Type-specific prevalence of high-risk human papillomavirus by cervical cytology and age: Data from the health check-ups of 7,014 Korean women. Obstet Gynecol Sci. 2013; 56:110–120.
Article14. Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, et al. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet Gynecol. 2013; 121:829–846.
Article15. Katki HA, Schiffman M, Castle PE, Fetterman B, Poitras NE, Lorey T, et al. Benchmarking CIN 3+ risk as the basis for incorporating HPV and Pap cotesting into cervical screening and management guidelines. J Low Genit Tract Dis. 2013; 17:S28–35.
Article16. Choi YD, Han CW, Chung WJ, Jung WW, Lee JS, Nam JH, et al. Analysis of HPV-other samples by performing HPV DNA sequencing. Korean J Pathol. 2009; 43:250–253.
Article17. Gharizadeh B, Oggionni M, Zheng B, Akom E, Pourmand N, Ahmadian A, et al. Type-specific multiple sequencing primers: a novel strategy for reliable and rapid genotyping of human papillomaviruses by pyrosequencing technology. J Mol Diagn. 2005; 7:198–205.18. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009; 10:321–322.
Article19. Schiffman MH, Bauer HM, Lorincz AT, Manos MM, Byrne JC, Glass AG, et al. Comparison of Southern blot hybridization and polymerase chain reaction methods for the detection of human papillomavirus DNA. J Clin Microbiol. 1991; 29:573–577.
Article20. Baay MF, Quint WG, Koudstaal J, Hollema H, Duk JM, Burger MP, et al. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol. 1996; 34:745–747.
Article21. Kado S, Kawamata Y, Shino Y, Kasai T, Kubota K, Iwasaki H, et al. Detection of human papillomaviruses in cervical neoplasias using multiple sets of generic polymerase chain reaction primers. Gynecol Oncol. 2001; 81:47–52.
Article22. Cho EJ, Do JH, Kim YS, Bae S, Ahn WS. Evaluation of a liquid bead array system for high-risk human papillomavirus detection and genotyping in comparison with Hybrid Capture II, DNA chip and sequencing methods. J Med Microbiol. 2011; 60:162–171.
Article23. Eklund C, Zhou T, Dillner J. WHO Human Papillomavirus Laboratory Network. Global proficiency study of human papillomavirus genotyping. J Clin Microbiol. 2010; 48:4147–4155.
Article24. Carvalho Nde O, del Castillo DM, Perone C, Januário JN, Melo VH, Brasileiro Filho G. Comparison of HPV genotyping by type-specific PCR and sequencing. Mem Inst Oswaldo Cruz. 2010; 105:73–78.25. Rao A, Young S, Erlich H, Boyle S, Krevolin M, Sun R, et al. Development and characterization of the cobas human papillomavirus test. J Clin Microbiol. 2013; 51:1478–1484.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of HPV-other Samples by Performing HPV DNA Sequencing
- Comparative Evaluation of the HPV28 Detection and HPV DNA Chip Test for Detecting and Genotyping Human Papillomaviruses
- Comparison of Four Human Papillomavirus Genotyping Methods: Next-generation Sequencing, INNO-LiPA, Electrochemical DNA Chip, and Nested-PCR
- Comparison of Clinical Efficacy between an HPV DNA Chip and a Hybrid-Capture II Assay in a Patient with Abnormal Colposcopic Findings
- Clinical Efficacy of HPV DNA Chip test in Human papillomavirus detection of Uterine Cervix