Yonsei Med J.
2018 Jan;59(1):141-147. 10.3349/ymj.2018.59.1.141.
microRNA-183 is Essential for Hair Cell Regeneration after Neomycin Injury in Zebrafish
- Affiliations
-
- 1Department of Otorhinolaryngology, Hallym University College of Medicine, Seoul, Korea.
- 2Department of Otorhinolaryngology, Yonsei University College of Medicine, Seoul, Korea. jychoi@yuhs.ac
- KMID: 2418861
- DOI: http://doi.org/10.3349/ymj.2018.59.1.141
Abstract
- PURPOSE
microRNAs (miRNAs) are non-coding RNAs composed of 20 to 22 nucleotides that regulate development and differentiation in various organs by silencing specific RNAs and regulating gene expression. In the present study, we show that the microRNA (miR)-183 cluster is upregulated during hair cell regeneration and that its inhibition reduces hair cell regeneration following neomycin-induced ototoxicity in zebrafish.
MATERIALS AND METHODS
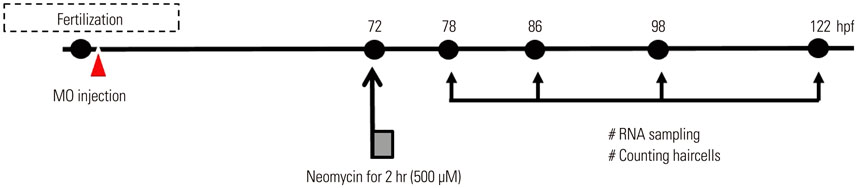
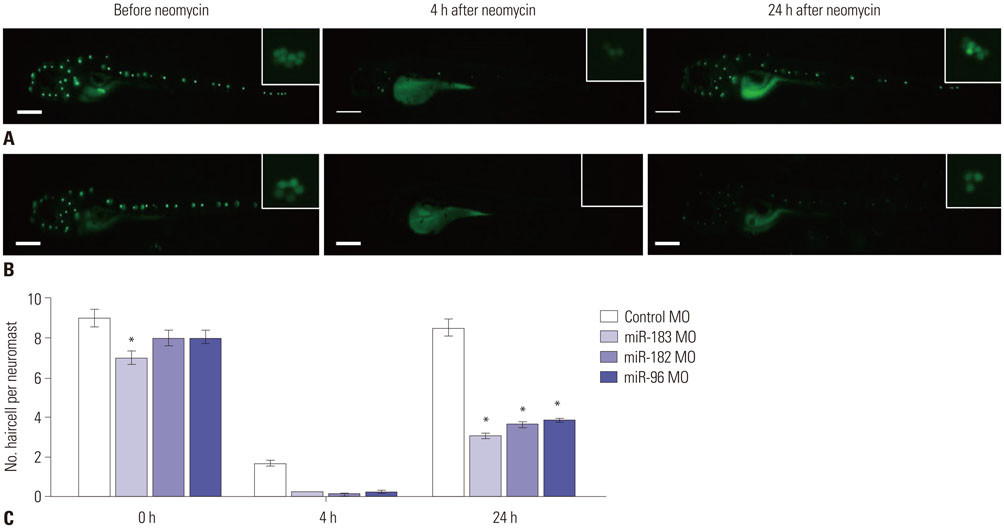
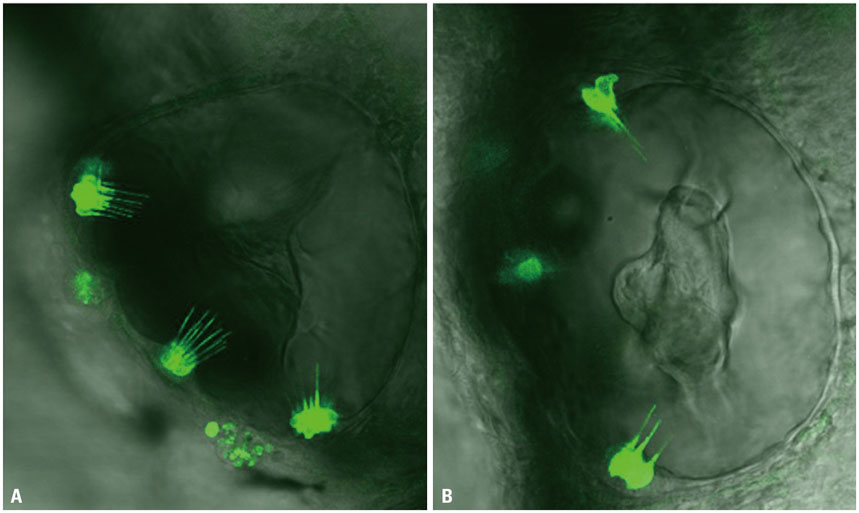
miRNA expression patterns after neomycin exposure were analyzed using microarray chips. Quantitative polymerase chain reaction was performed to validate miR-183 cluster expression patterns following neomycin exposure (500 µM for 2 h). After injection of an antisense morpholino (MO) to miR-183 (MO-183) immediately after fertilization, hair cell regeneration after neomycin exposure in neuromast cells was evaluated by fluorescent staining (YO-PRO1). The MO-183 effect also was assessed in transgenic zebrafish larvae expressing green fluorescent protein (GFP) in inner ear hair cells.
RESULTS
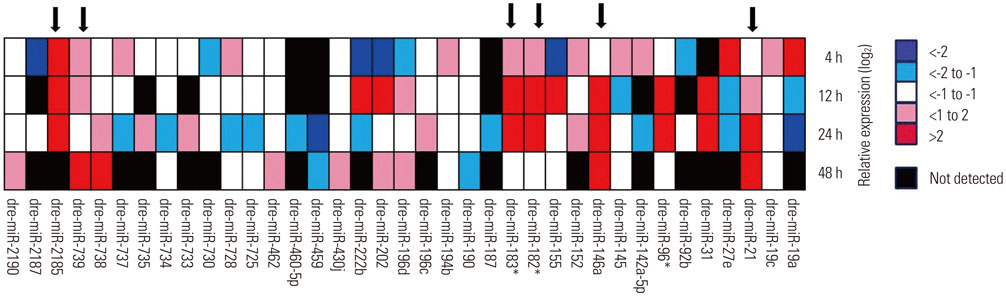
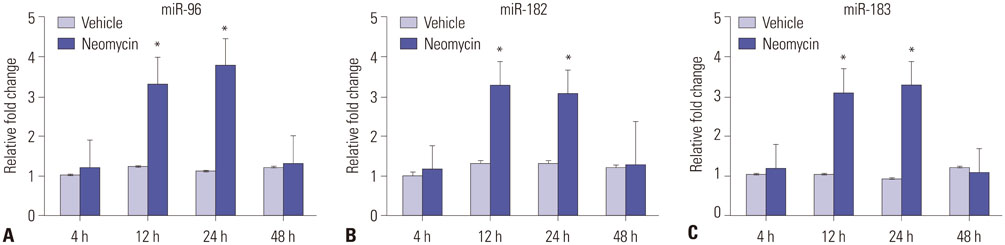
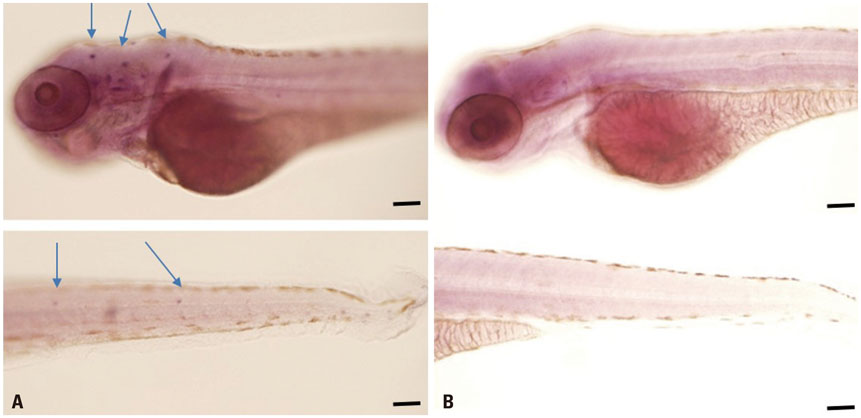
Microarray analysis clearly showed that the miR-183 cluster (miR-96, miR-182, and miR-183) was upregulated after neomycin treatment. We also confirmed upregulated expression of the miR-183 cluster during hair cell regeneration after neomycin-induced ototoxicity. miR-183 inhibition using MO-183 reduced hair cell regeneration in both wild-type and GFP transgenic zebrafish larvae.
CONCLUSION
Our work demonstrates that the miR-183 cluster is essential for the regeneration of hair cells following ototoxic injury in zebrafish larvae. Therefore, regulation of the miR-183 cluster can be a novel target for stimulation of hair cell regeneration.
Keyword
MeSH Terms
-
Animals
Animals, Genetically Modified
Cell Count
Gene Expression Profiling
Gene Expression Regulation/drug effects
Gene Knockdown Techniques
Green Fluorescent Proteins/metabolism
Hair Cells, Auditory/drug effects/*physiology
Larva/drug effects/genetics
MicroRNAs/genetics/*metabolism
Morpholinos/pharmacology
Neomycin/toxicity
Regeneration/drug effects/*genetics
Zebrafish/*genetics
MicroRNAs
Morpholinos
Neomycin
Green Fluorescent Proteins
Figure
Reference
-
1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297.2. Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005; 309:310–311.
Article3. Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006; 1111:95–104.
Article4. Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010; 30:3254–3263.
Article5. Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, et al. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011; 240:808–819.
Article6. Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009; 106:7915–7920.
Article7. Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009; 41:614–618.
Article8. Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009; 41:609–613.
Article9. King BL, Yin VP. A conserved microRNA regulatory circuit is differentially controlled during limb/appendage regeneration. PLoS One. 2016; 11:e0157106.
Article10. Gurha P. MicroRNAs in cardiovascular disease. Curr Opin Cardiol. 2016; 31:249–254.
Article11. Yi PS, Zhang M, Xu MQ. Role of microRNA in liver regeneration. Hepatobiliary Pancreat Dis Int. 2016; 15:141–146.
Article12. Li Y, Li A, Wu J, He Y, Yu H, Chai R, et al. MiR-182-5p protects inner ear hair cells from cisplatin-induced apoptosis by inhibiting FOXO3a. Cell Death Dis. 2016; 7:e2362.
Article13. Tsonis PA, Call MK, Grogg MW, Sartor MA, Taylor RR, Forge A, et al. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem Biophys Res Commun. 2007; 362:940–945.
Article14. Xiao T, Roeser T, Staub W, Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005; 132:2955–2967.
Article15. Moon IS, So JH, Jung YM, Lee WS, Kim EY, Choi JH, et al. Fucoidan promotes mechanosensory hair cell regeneration following amino glycoside-induced cell death. Hear Res. 2011; 282:236–242.
Article16. Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, et al. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010; 5:e14302.
Article17. Yu H, Liu Y, Bai L, Kijlstra A, Yang P. Predisposition to Behçet's disease and VKH syndrome by genetic variants of miR-182. J Mol Med (Berl). 2014; 92:961–967.
Article18. Sánchez-Mora C, Ramos-Quiroga JA, Garcia-Martínez I, Fernàndez-Castillo N, Bosch R, Richarte V, et al. Evaluation of single nucleotide polymorphisms in the miR-183-96-182 cluster in adulthood attention-deficit and hyperactivity disorder (ADHD) and substance use disorders (SUDs). Eur Neuropsychopharmacol. 2013; 23:1463–1473.
Article19. Wang G, Wang S, Li C. MiR-183 overexpression inhibits tumorigenesis and enhances DDP-induced cytotoxicity by targeting MTA1 in nasopharyngeal carcinoma. Tumour Biol. 2017; 39:1010428317703825.
Article20. Lin HC, Liu SY, Yen EY, Li TK, Lai IR. microRNA-183 mediates protective postconditioning of the liver by repressing Apaf-1. Antioxid Redox Signal. 2017; 26:583–597.
Article21. Frucht CS, Uduman M, Duke JL, Kleinstein SH, Santos-Sacchi J, Navaratnam DS. Gene expression analysis of forskolin treated basilar papillae identifies microRNA181a as a mediator of proliferation. PLoS One. 2010; 5:e11502.
Article22. Frucht CS, Santos-Sacchi J, Navaratnam DS. MicroRNA181a plays a key role in hair cell regeneration in the avian auditory epithelium. Neurosci Lett. 2011; 493:44–48.
Article23. Wang Z, Liu Y, Han N, Chen X, Yu W, Zhang W, et al. Profiles of oxidative stress-related microRNA and mRNA expression in auditory cells. Brain Res. 2010; 1346:14–25.
Article24. Yu L, Tang H, Jiang XH, Tsang LL, Chung YW, Chan HC. Involvement of calpain-I and microRNA34 in kanamycin-induced apoptosis of inner ear cells. Cell Biol Int. 2010; 34:1219–1225.
Article25. Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 2006; 27:1–19.
Article26. Jiang H, Talaska AE, Schacht J, Sha SH. Oxidative imbalance in the aging inner ear. Neurobiol Aging. 2007; 28:1605–1612.
Article27. Patel M, Cai Q, Ding D, Salvi R, Hu Z, Hu BH. The miR-183/Taok1 target pair is implicated in cochlear responses to acoustic trauma. PLoS One. 2013; 8:e58471.
Article28. Cardano M, Diaferia GR, Cattaneo M, Dessì SS, Long Q, Conti L, et al. mSEL-1L (Suppressor/enhancer Lin12-like) protein levels influence murine neural stem cell self-renewal and lineage commitment. J Biol Chem. 2011; 286:18708–18719.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Protective Role of Trimetazidine Against Neomycin-induced Hair Cell Damage in Zebrafish
- MicroRNA-183 Family in Inner Ear: Hair Cell Development and Deafness
- Hair Cell Regeneration: From Animals to Humans
- Notch Signaling Controls Oligodendrocyte Regeneration in the Injured Telencephalon of Adult Zebrafish
- Zebrafish and Mycobacterial infection