Yonsei Med J.
2018 Jan;59(1):13-19. 10.3349/ymj.2018.59.1.13.
Detection of Rare Mutations in EGFR-ARMS-PCR-Negative Lung Adenocarcinoma by Sanger Sequencing
- Affiliations
-
- 1Guangzhou Institute of Respiratory Disease, Guangzhou, China. drzhangjx@126.com
- 2Department of Internal Medicine, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
- 3Department of Pulmonary Medicine, The Brain Hospital of Guangxi Zhuang Autonomous Region, Liuzhou, China.
- 4Department of Biomedical Engineering, University of Minnesota, Twin Cities, Minneapolis, USA.
- 5Guangzhou Life Technologies Daan Diagnostics Co., Ltd., Guangzhou, China. chen@mendel-genes.com
- 6Mendel Genes, Inc., Manhattan Beach, CA, USA.
- 7Department of Health Care, Maternal and Child Health Hospital of Haizhu District, Guangzhou, China.
- KMID: 2418843
- DOI: http://doi.org/10.3349/ymj.2018.59.1.13
Abstract
- PURPOSE
This study aimed to identify potential epidermal growth factor receptor (EGFR) gene mutations in non-small cell lung cancer that went undetected by amplification refractory mutation system-Scorpion real-time PCR (ARMS-PCR).
MATERIALS AND METHODS
A total of 200 specimens were obtained from the First Affiliated Hospital of Guangzhou Medical University from August 2014 to August 2015. In total, 100 ARMS-negative and 100 ARMS-positive specimens were evaluated for EGFR gene mutations by Sanger sequencing. The methodology and sensitivity of each method and the outcomes of EGFR-tyrosine kinase inhibitor (TKI) therapy were analyzed.
RESULTS
Among the 100 ARMS-PCR-positive samples, 90 were positive by Sanger sequencing, while 10 cases were considered negative, because the mutation abundance was less than 10%. Among the 100 negative cases, three were positive for a rare EGFR mutation by Sanger sequencing. In the curative effect analysis of EGFR-TKIs, the progression-free survival (PFS) analysis based on ARMS and Sanger sequencing results showed no difference. However, the PFS of patients with a high abundance of EGFR mutation was 12.4 months [95% confidence interval (CI), 11.6−12.4 months], which was significantly higher than that of patients with a low abundance of mutations detected by Sanger sequencing (95% CI, 10.7−11.3 months) (p < 0.001).
CONCLUSION
The ARMS method demonstrated higher sensitivity than Sanger sequencing, but was prone to missing mutations due to primer design. Sanger sequencing was able to detect rare EGFR mutations and deemed applicable for confirming EGFR status. A clinical trial evaluating the efficacy of EGFR-TKIs in patients with rare EGFR mutations is needed.
MeSH Terms
-
Adenocarcinoma/*genetics/pathology
Aged
Aged, 80 and over
Animals
Base Sequence
Disease-Free Survival
Female
Humans
Lung Neoplasms/*genetics/pathology
Male
Middle Aged
Mutation/*genetics
Mutation Rate
Real-Time Polymerase Chain Reaction/*methods
Receptor, Epidermal Growth Factor/*genetics
Sequence Analysis, DNA/*methods
Treatment Outcome
Receptor, Epidermal Growth Factor
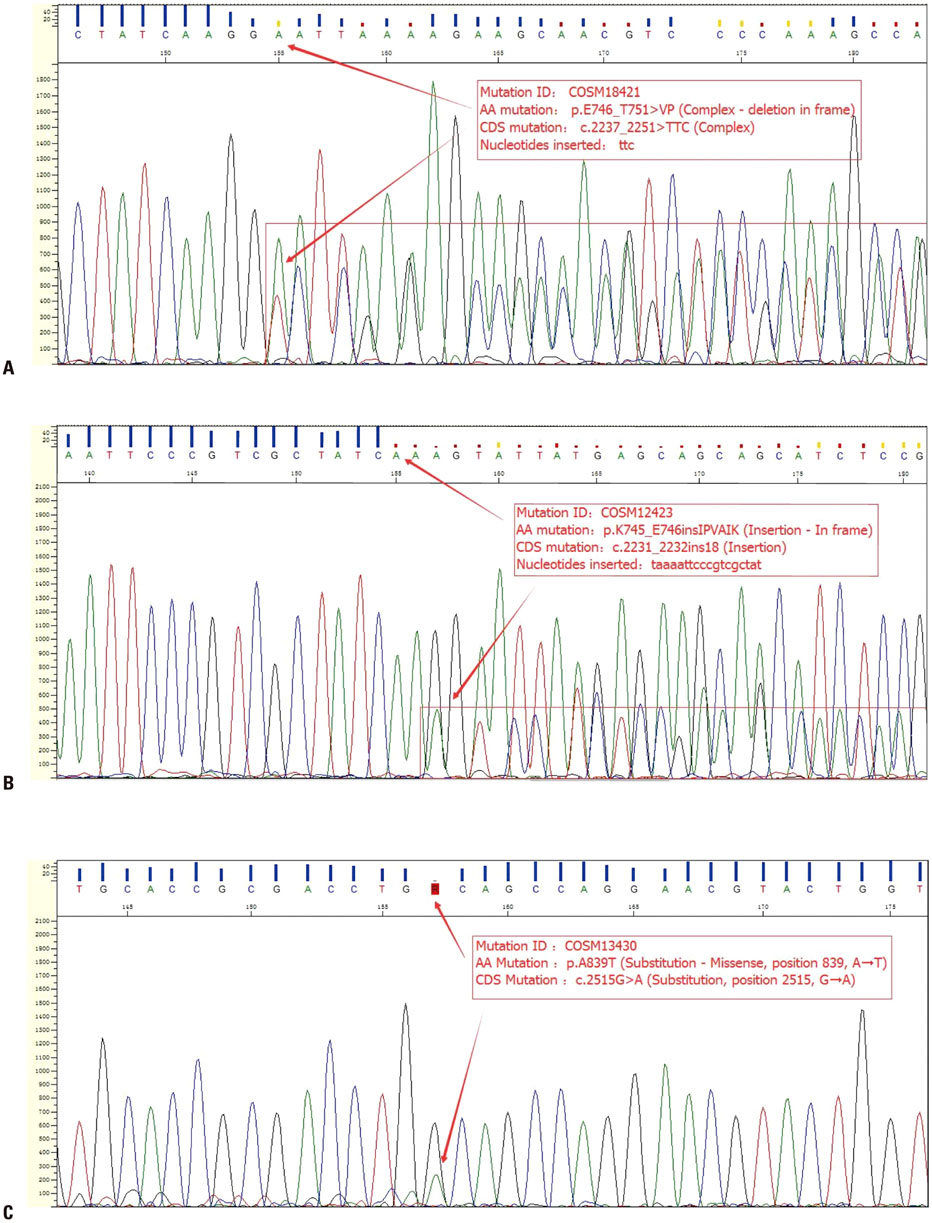
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65:5–29.
Article2. Tan DS, Mok TS, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J Clin Oncol. 2016; 34:91–101.
Article3. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010; 362:2380–2388.
Article4. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive nonsmall-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012; 13:239–246.5. Lee JY, Sun JM, Lim SH, Kim HS, Yoo KH, Jung KS, et al. A phase Ib/II study of afatinib in combination with nimotuzumab in non-small cell lung cancer patients with acquired resistance to gefitinib or erlotinib. Clin Cancer Res. 2016; 22:2139–2145.
Article6. Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015; 372:1689–1699.
Article7. Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013; 66:1065–1069.
Article8. Liu J, Zhao R, Zhang J, Zhang J. ARMS for EGFR mutation analysis of cytologic and corresponding lung adenocarcinoma histologic specimens. J Cancer Res Clin Oncol. 2015; 141:221–227.
Article9. Min KW, Kim WS, Jang SJ, Choi YD, Chang S, Jung SH, et al. Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study. QJM. 2016; 109:167–173.
Article10. Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008; 8:142.
Article11. Ross JS, Cronin M. Whole cancer genome sequencing by nextgeneration methods. Am J Clin Pathol. 2011; 136:527–539.
Article12. Hu N, Wang G, Wu YH, Chen SF, Liu GD, Chen C, et al. LDA-SVM-based EGFR mutation model for NSCLC brain metastases: an observational study. Medicine (Baltimore). 2015; 94:e375.13. Zhao ZR, Wang JF, Lin YB, Wang F, Fu S, Zhang SL, et al. Mutation abundance affects the efficacy of EGFR tyrosine kinase inhibitor readministration in non-small-cell lung cancer with acquired resistance. Med Oncol. 2014; 31:810.
Article14. Shaozhang Z, Ming Z, Haiyan P, Aiping Z, Qitao Y, Xiangqun S. Comparison of ARMS and direct sequencing for detection of EGFR mutation and prediction of EGFR-TKI efficacy between surgery and biopsy tumor tissues in NSCLC patients. Med Oncol. 2014; 31:926.
Article15. Li Y, Zhang Q. A Weibull multi-state model for the dependence of progression-free survival and overall survival. Stat Med. 2015; 34:2497–2513.
Article16. Zhou Q, Zhang XC, Chen ZH, Yin XL, Yang JJ, Xu CR, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol. 2011; 29:3316–3321.
Article17. Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012; 10:1236–1271.
Article18. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004; 64:8919–8923.
Article19. Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005; 97:339–346.
Article20. Sasaki H, Shimizu S, Endo K, Takada M, Kawahara M, Tanaka H, et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int J Cancer. 2006; 118:180–184.
Article21. Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005; 11:1167–1173.22. Yang X, Yang K, Kuang K. The efficacy and safety of EGFR inhibitor monotherapy in non-small cell lung cancer: a systematic review. Curr Oncol Rep. 2014; 16:390.
Article23. Liu Y, Liu B, Li XY, Li JJ, Qin HF, Tang CH, et al. A comparison of ARMS and direct sequencing for EGFR mutation analysis and tyrosine kinase inhibitors treatment prediction in body fluid samples of non-small-cell lung cancer patients. J Exp Clin Cancer Res. 2011; 30:111.
Article24. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lungcancer cells. N Engl J Med. 2008; 359:366–377.
Article25. Yang JC, Sequist LV, Zhou C, Schuler M, Geater SL, Mok T, et al. Effect of dose adjustment on the safety and efficacy of afatinib for EGFR mutation-positive lung adenocarcinoma: post hoc analyses of the randomized LUX-Lung 3 and 6 trials. Ann Oncol. 2016; 27:2103–2110.
Article26. Voon PJ, Tsui DW, Rosenfeld N, Chin TM. EGFR exon 20 insertion A763-Y764insFQEA and response to erlotinib–Letter. Mol Cancer Ther. 2013; 12:2614–2615.
Article27. Baek JH, Sun JM, Min YJ, Cho EK, Cho BC, Kim JH, et al. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small cell lung cancer except both exon 19 deletion and exon 21 L858R: a retrospective analysis in Korea. Lung Cancer. 2015; 87:148–154.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
- Screening of Epidermal Growth Factor Receptor Gene Mutation in Non-Small Cell Lung Cancer Using a PCR-Based Enzymatic Digestion Method
- A Case of Patient with Lung Adenocarcinoma with Double Rare EGFR Mutation of G719C and L861Q
- KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay
- Screening PCR Versus Sanger Sequencing: Detection of CALR Mutations in Patients With Thrombocytosis