Yonsei Med J.
2018 Aug;59(6):760-768. 10.3349/ymj.2018.59.6.760.
Exploring the Key Genes and Pathways of Osteoarthritis in Knee Cartilage in a Rat Model Using Gene Expression Profiling
- Affiliations
-
- 1Department of Joint and Sport Medicine, Tianjin Union Medical Center, Tianjin, China. tmqjoint@126.com
- 2Nankai Clinical College, Tianjin Medical University, Tianjin, China.
- KMID: 2415535
- DOI: http://doi.org/10.3349/ymj.2018.59.6.760
Abstract
- PURPOSE
To compare differentially expressed genes (DEGs) mediating osteoarthritis (OA) in knee cartilage and in normal knee cartilage in a rat model of OA and to identify their impact on molecular pathways associated with OA.
MATERIALS AND METHODS
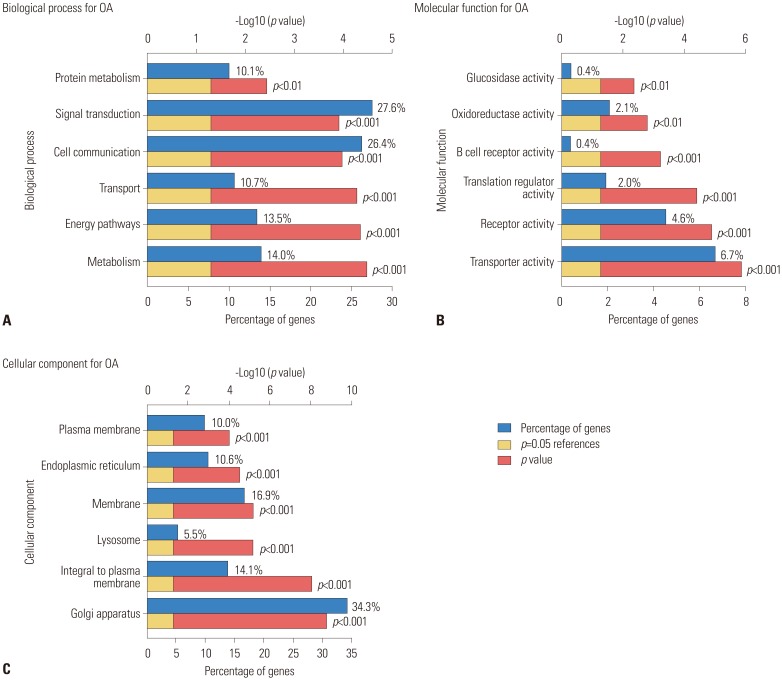
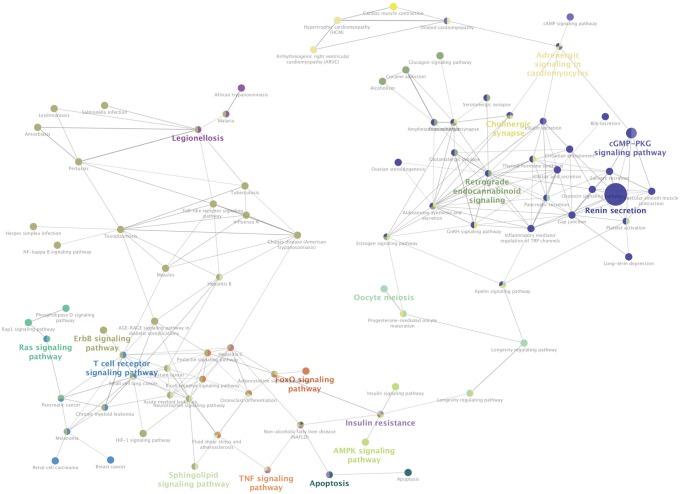
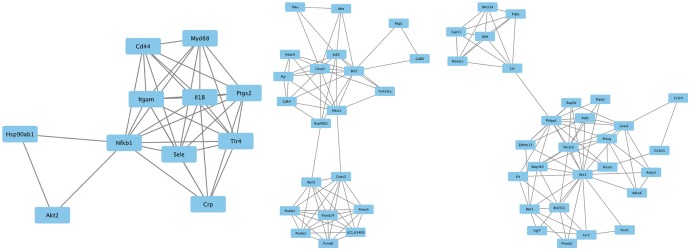
A gene expression profile was downloaded from the Gene Expression Omnibus database. Analysis of DEGs was carried out using GEO2R. Enrichment analyses were performed on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway using the Search Tool for the Retrieval of Interacting Genes database (http://www.string-db.org/). Subsequently, the regulatory interaction network of OA-associated genes was visualized using Cytoscape software (version 3.4.0; www.cytoscape.org).
RESULTS
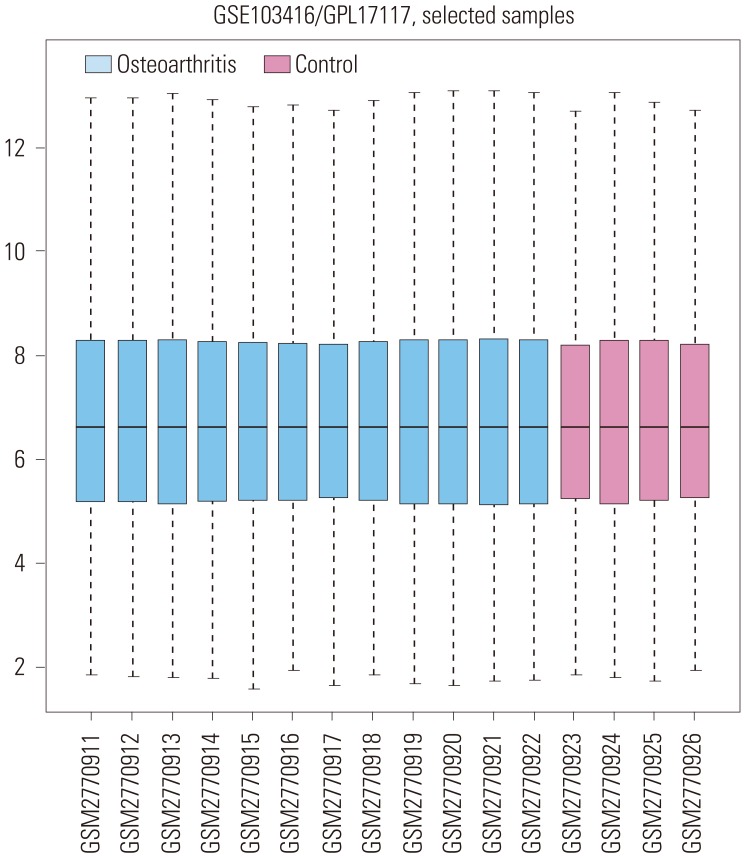
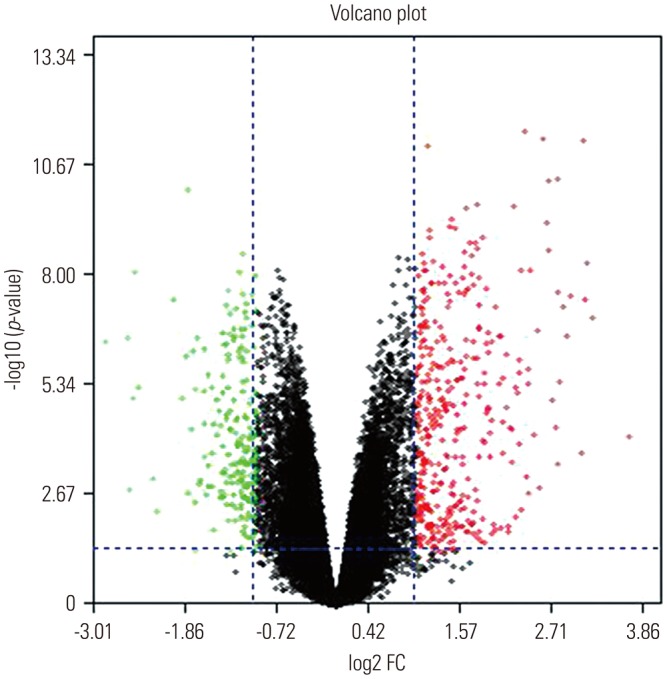
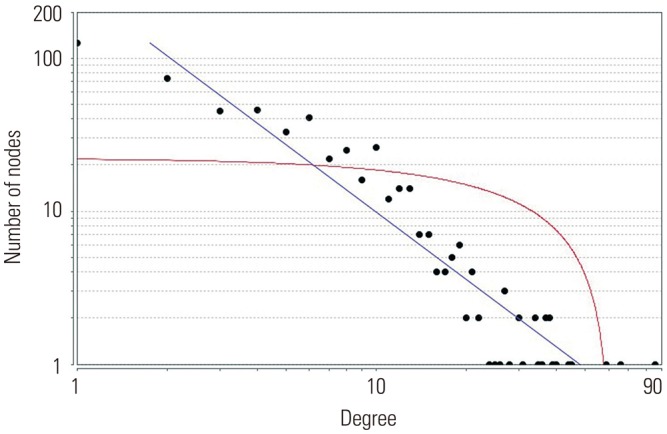
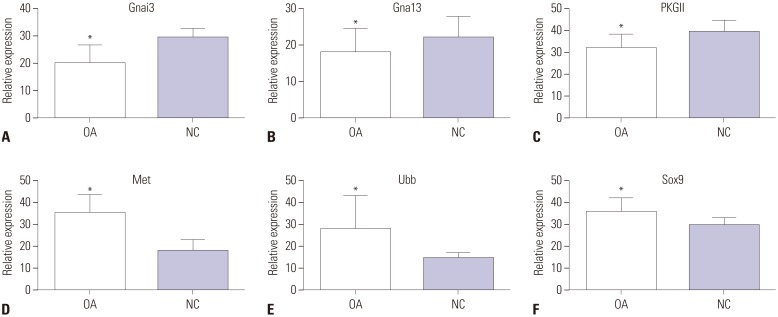
In the gene expression profile GSE103416, a total of 99 DEGs were identified. Among them, 76 DEGs (76.77%) were overexpressed, and the remaining 23 DEGs (23.23%) were underexpressed. GO and pathway enrichment analyses of target genes were performed. Using gene-gene interaction network analysis, relevant core genes, including MET, UBB, GNAI3, and GNA13, were shown to hold a potential relationship with the development of OA in cartilage. Using quantitative real-time PCR, the Gna13/cGMP-PKG signaling pathway was identified as a potential research target for therapy and for further understanding the development of OA.
CONCLUSION
The results of the present study provide a comprehensive understanding of the roles of DEGs in knee cartilage in relation to the development of OA.
MeSH Terms
Figure
Reference
-
1. Hussain SM, Neilly DW, Baliga S, Patil S, Meek R. Knee osteoarthritis: a review of management options. Scott Med J. 2016; 61:7–16. PMID: 27330013.
Article2. Ayral X, Pickering EH, Woodworth TG, Mackillop N, Dougados M. Synovitis: a potential predictive factor of structural progression of medial tibiofemoral knee osteoarthritis -- results of a 1 year longitudinal arthroscopic study in 422 patients. Osteoarthritis Cartilage. 2005; 13:361–367. PMID: 15882559.
Article3. Lohmander LS, Ionescu M, Jugessur H, Poole AR. Changes in joint cartilage aggrecan after knee injury and in osteoarthritis. Arthritis Rheum. 1999; 42:534–544. PMID: 10088777.
Article4. Brandt KD, Dieppe P, Radin EL. Etiopathogenesis of osteoarthritis. Rheum Dis Clin North Am. 2008; 34:531–559. PMID: 18687271.
Article5. Lotz MK, Kraus VB. New developments in osteoarthritis. Posttraumatic osteoarthritis: pathogenesis and pharmacological treatment options. Arthritis Res Ther. 2010; 12:211. PMID: 20602810.
Article6. Martin I, Jakob M, Schäfer D, Dick W, Spagnoli G, Heberer M. Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthritis Cartilage. 2001; 9:112–118. PMID: 11237658.
Article7. Felson DT, Neogi T. Osteoarthritis: is it a disease of cartilage or of bone? Arthritis Rheum. 2004; 50:341–344. PMID: 14872473.
Article8. Rogers J, Shepstone L, Dieppe P. Is osteoarthritis a systemic disorder of bone? Arthritis Rheum. 2004; 50:452–457. PMID: 14872487.
Article9. Xu L, Sun C, Zhang S, Xu X, Zhai L, Wang Y, et al. Sam68 promotes NF-κB activation and apoptosis signaling in articular chondrocytes during osteoarthritis. Inflamm Res. 2015; 64:895–902. PMID: 26350037.
Article10. He W, Cheng Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018; 97:607–615. PMID: 29101804.
Article11. Li X, Li J, Cheng K, Lin Q, Wang D, Zhang H, et al. Effect of low-intensity pulsed ultrasound on MMP-13 and MAPKs signaling pathway in rabbit knee osteoarthritis. Cell Biochem Biophys. 2011; 61:427–434. PMID: 21567132.
Article12. Xue H, Tu Y, Ma T, Liu X, Wen T, Cai M, et al. Lactoferrin inhibits IL-1β-induced chondrocyte apoptosis through AKT1-induced CREB1 activation. Cell Physiol Biochem. 2015; 36:2456–2465. PMID: 26279447.
Article13. Bradley EW, Carpio LR, Newton AC, Westendorf JJ. Deletion of the PH-domain and leucine-rich repeat protein phosphatase 1 (Phlpp1) increases fibroblast growth factor (Fgf) 18 expression and promotes chondrocyte proliferation. J Biol Chem. 2015; 290:16272–16280. PMID: 25953896.
Article14. Bradley EW, Carpio LR, McGee-Lawrence ME, Castillejo Becerra C, Amanatullah DF, Ta LE, et al. Phlpp1 facilitates post-traumatic osteoarthritis and is induced by inflammation and promoter demethylation in human osteoarthritis. Osteoarthritis Cartilage. 2016; 24:1021–1028. PMID: 26746148.
Article15. Zhou Y, Liu SQ, Yu L, He B, Wu SH, Zhao Q, et al. Berberine prevents nitric oxide-induced rat chondrocyte apoptosis and cartilage degeneration in a rat osteoarthritis model via AMPK and p38 MAPK signaling. Apoptosis. 2015; 20:1187–1199. PMID: 26184498.
Article16. Culley KL, Hui W, Barter MJ, Davidson RK, Swingler TE, Destrument AP, et al. Class I histone deacetylase inhibition modulates metalloproteinase expression and blocks cytokine-induced cartilage degradation. Arthritis Rheum. 2013; 65:1822–1830. PMID: 23575963.
Article17. Zhang ZM, Shen C, Li H, Fan Q, Ding J, Jin FC, et al. Leptin induces the apoptosis of chondrocytes in an in vitro model of osteoarthritis via the JAK2STAT3 signaling pathway. Mol Med Rep. 2016; 13:3684–3690. PMID: 26936086.
Article18. Valdes AM, Van Oene M, Hart DJ, Surdulescu GL, Loughlin J, Doherty M, et al. Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis Rheum. 2006; 54:533–539. PMID: 16453284.
Article19. Gómez R, Villalvilla A, Largo R, Gualillo O, Herrero-Beaumont G. TLR4 signalling in osteoarthritis--finding targets for candidate DMOADs. Nat Rev Rheumatol. 2015; 11:159–170. PMID: 25512010.
Article20. Schneider EM, Du W, Fiedler J, Högel J, Günther KP, Brenner H, et al. The (−765 G→C) promoter variant of the COX-2/PTGS2 gene is associated with a lower risk for end-stage hip and knee osteoarthritis. Ann Rheum Dis. 2011; 70:1458–1460. PMID: 20378913.
Article21. Zhu X, Yang S, Lin W, Wang L, Ying J, Ding Y, et al. Roles of cell cyle regulators cyclin D1, CDK4, and p53 in knee osteoarthritis. Genet Test Mol Biomarkers. 2016; 20:529–534. PMID: 27391794.
Article22. Feng Z, Lian KJ. Identification of genes and pathways associated with osteoarthritis by bioinformatics analyses. Eur Rev Med Pharmacol Sci. 2015; 19:736–744. PMID: 25807424.23. Tian H. Detection of differentially expressed genes involved in osteoarthritis pathology. J Orthop Surg Res. 2018; 13:49. PMID: 29514675.
Article24. Mi B, Liu G, Zhou W, Lv H, Liu Y, Liu J. Identification of genes and pathways in the synovia of women with osteoarthritis by bioinformatics analysis. Mol Med Rep. 2018; 17:4467–4473. PMID: 29344651.
Article25. Li M, Zhi L, Zhang Z, Bian W, Qiu Y. Identification of potential target genes associated with the pathogenesis of osteoarthritis using microarray based analysis. Mol Med Rep. 2017; 16:2799–2806. PMID: 28714028.
Article26. Zhang X, Yuan Z, Cui S. Identifying candidate genes involved in osteoarthritis through bioinformatics analysis. Clin Exp Rheumatol. 2016; 34:282–290. PMID: 26968041.27. Sun J, Yan B, Yin W, Zhang X. Identification of genes associated with osteoarthritis by microarray analysis. Mol Med Rep. 2015; 12:5211–5216. PMID: 26151199.
Article28. Wang Q, Li Y, Zhang Z, Fang Y, Li X, Sun Y, et al. Bioinformatics analysis of gene expression profiles of osteoarthritis. Acta Histochem. 2015; 117:40–46. PMID: 25466988.
Article29. Park KH, Park B, Yoon DS, Kwon SH, Shin DM, Lee JW, et al. Zinc inhibits osteoclast differentiation by suppression of Ca2+-calcineurin-NFATc1 signaling pathway. Cell Commun Signal. 2013; 11:74. PMID: 24088289.
Article30. Chikuda H, Kugimiya F, Hoshi K, Ikeda T, Ogasawara T, Shimoaka T, et al. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004; 18:2418–2429. PMID: 15466490.
Article31. Kawasaki Y, Kugimiya F, Chikuda H, Kamekura S, Ikeda T, Kawamura N, et al. Phosphorylation of GSK-3beta by cGMP-dependent protein kinase II promotes hypertrophic differentiation of murine chondrocytes. J Clin Invest. 2008; 118:2506–2515. PMID: 18551195.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Exploring molecular mechanisms underlying the pathophysiological association between knee osteoarthritis and sarcopenia
- Comparative co-expression analysis of RNA-Seq transcriptome revealing key genes, miRNA and transcription factor in distinct metabolic pathways in diabetic nerve, eye, and kidney disease
- Gene Expression Profiling by Microarray during Tooth Development of Rats
- Effect of Displacement and Morphological Change of Medial Meniscus on Early Osteoarthritis of the Knee
- Arthroscopic Management of Osteoarthritic Knee