Transl Clin Pharmacol.
2017 Mar;25(1):5-9. 10.12793/tcp.2017.25.1.5.
Forensic science meets clinical pharmacology: pharmacokinetic model based estimation of alcohol concentration of a defendant as requested by a local prosecutor's office
- Affiliations
-
- 1Department of Clinical Pharmacology and Therapeutics, Asan Medical Center, Ulsan University College of Medicine, Seoul 05505, Republic of Korea. ksbae@amc.seoul.kr
- 2Center of International Cooperation, Korean Institute of Criminology, Seoul 06764, Republic of Korea.
- KMID: 2411412
- DOI: http://doi.org/10.12793/tcp.2017.25.1.5
Abstract
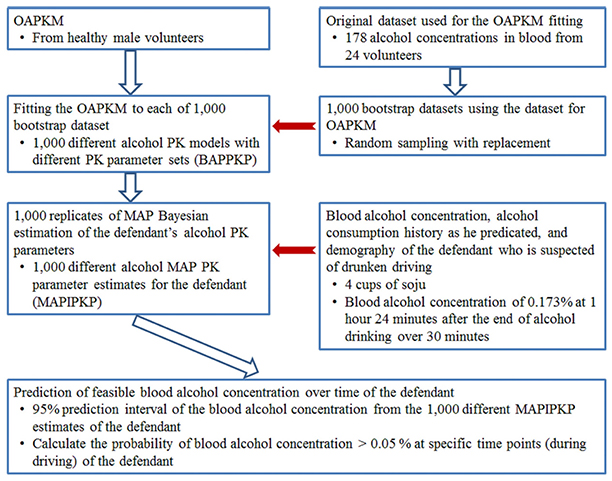
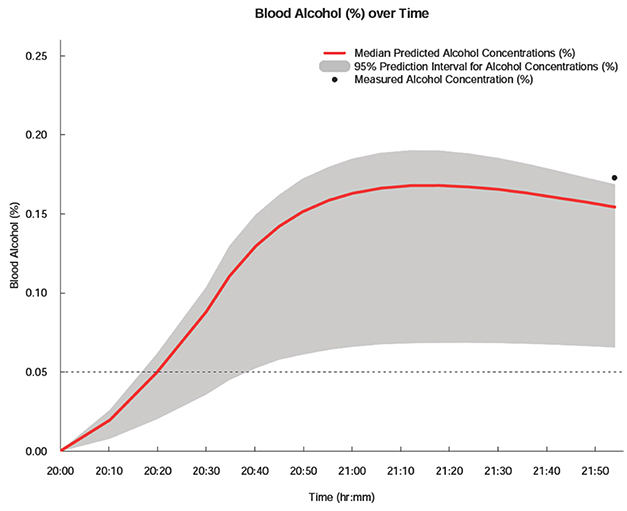
- Drunk driving is a serious social problem. We estimated the blood alcohol concentration of a defendant on the request of local prosecutor's office in Korea. Based on the defendant's history, and a previously constructed pharmacokinetic model for alcohol, we estimated the possible alcohol concentration over time during his driving using a Bayesian method implemented in NONMEM®. To ensure generalizability and to take the parameter uncertainty of the alcohol pharmacokinetic models into account, a non-parametric bootstrap with 1,000 replicates was applied to the Bayesian estimations. The current analysis enabled the prediction of the defendant's possible blood alcohol concentrations over time with a 95% prediction interval. The results showed a high probability that the alcohol concentration was ≥ 0.05% during driving. The current estimation of the alcohol concentration during driving by the Bayesian method could be used as scientific evidence during court trials.
Keyword
MeSH Terms
Figure
Reference
-
1. KoROAD webzine. Accessed November 25 2016. Retrieved from http://news.koroad.or.kr/articleview.php?idx=112.2. Bruno R, Iliadis A, Botta A, Mariotti B, Jullien G, Cano JP. Pharmacokinetic study of ethanol after oral administration: a new approach to enzymatic elimination. Int J Clin Pharmacol Ther Toxicol. 1983; 21:363–369.3. Gotta V, Widmer N, Montemurro M, Leyvraz S, Haouala A, Decosterd LA, et al. Therapeutic drug monitoring of imatinib: Bayesian and alternative methods to predict trough levels. Clin Pharmacokinet. 2012; 51:187–201. DOI: 10.2165/11596990-000000000-00000.4. Saint-Marcoux F, Marquet P, Jacqz-Aigrain E, Bernard N, Thiry P, Le Meur Y, et al. Patient characteristics influencing ciclosporin pharmacokinetics and accurate Bayesian estimation of ciclosporin exposure in heart, lung and kidney transplant patients. Clin Pharmacokinet. 2006; 45:905–922.
Article5. Parke J, Holford NH, Charles BG. A procedure for generating bootstrap samples for the validation of nonlinear mixed-effects population models. Comput Methods Programs Biomed. 1999; 59:19–29.
Article6. Parke J, Charles BG. Factors affecting oral cyclosporin disposition after heart transplantation: bootstrap validation of a population pharmacokinetic model. Eur J Clin Pharmacol. 2000; 56:481–487.
Article7. Holford NH, Kirkpatrick C, Duffull D. NONMEM Termination status is not an important indicator of the quality of bootstrap parameter estimates. PAGE 2006;Abstr 992. Accessed Novermber 25 2016. http://www.page-meeting.org/default.asp?abstract=992.8. Gastonguay MR, El-Tahtawy A. Effect of NONMEM minimization status and number of replicates on bootstrap parameter distributions for population pharmacokinetic models: a case study. Clin Pharmacol Ther. 2005; 77:P2–P2.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Estimation of Fentanyl Concentrations Based on a Pharmacokinetic Model
- Likelihood interval for nonlinear regression
- A semi-compartmental model describing the pharmacokinetic-pharmacodynamic relationship
- PKconverter: R package to convert the pharmacokinetic parameters
- A Detailed Analysis of Alcohol Pharmacokinetics in Healthy Korean Men