Cancer Res Treat.
2017 Oct;49(4):1077-1087. 10.4143/crt.2016.301.
Identification of Diverse Adenosine-to-Inosine RNA Editing Subtypes in Colorectal Cancer
- Affiliations
-
- 1Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University College of Medicine, Seoul, Korea. kimty@snu.ac.kr
- 2Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea.
- 3Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea.
- KMID: 2394826
- DOI: http://doi.org/10.4143/crt.2016.301
Abstract
- PURPOSE
RNA editing generates protein diversity by altering RNA sequences in coding regions without changing the overall DNA sequence. Adenosine-to-inosine (A-to-I) RNA editing events have recently been reported in some types of cancer, but they are rare in human colorectal cancer (CRC). Therefore, this study was conducted to identify diverse RNA editing in CRC.
MATERIALS AND METHODS
We compared transcriptome data of 39 CRC samples and paired adjacent tissues from The Cancer Genome Atlas database to identify RNA editing patterns in CRC, focusing on canonical A-to-I RNA edits in coding sequence regions. We investigated nonsynonymous RNA editing patterns by comparing tumor and normal tissue transcriptome data.
RESULTS
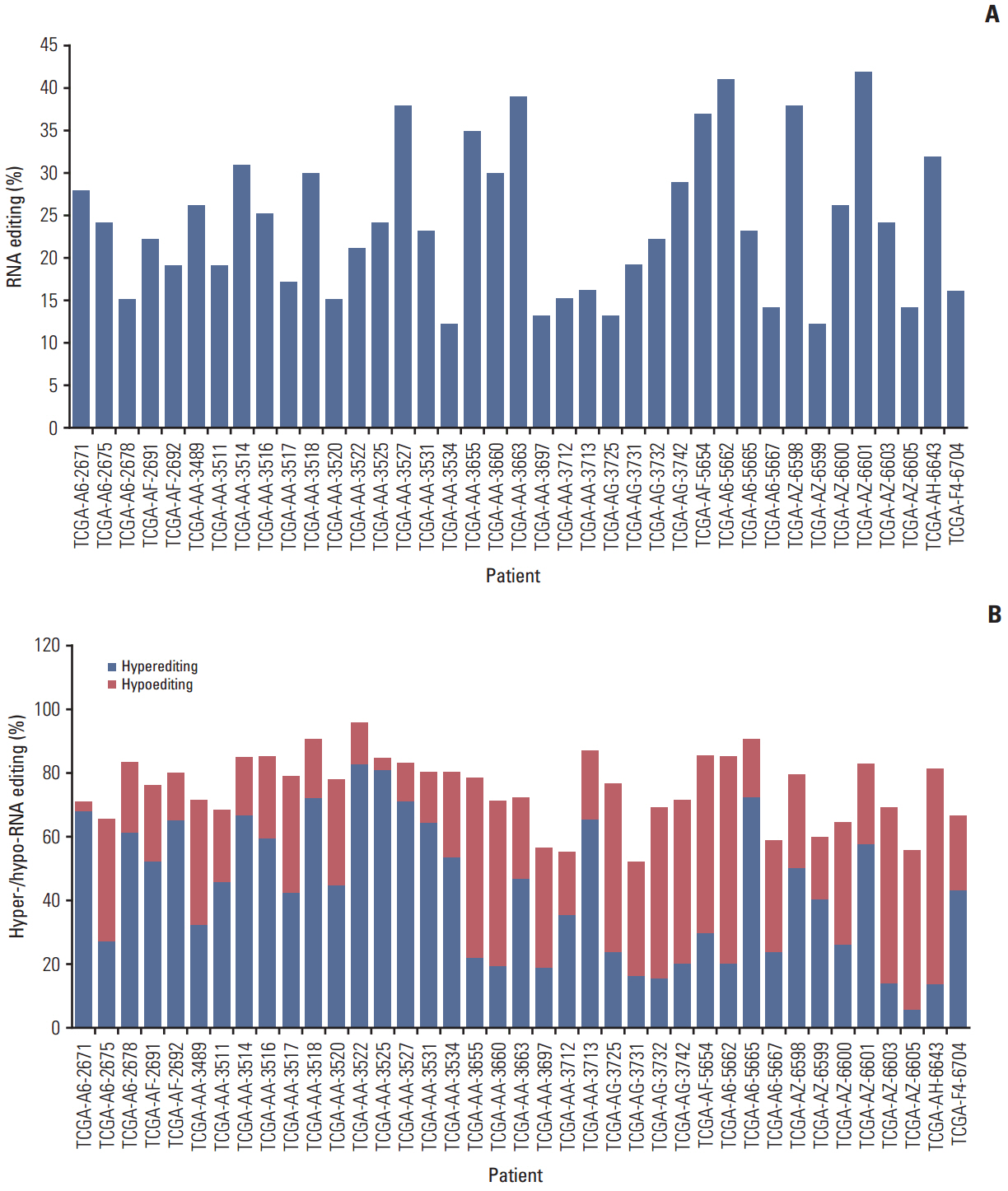
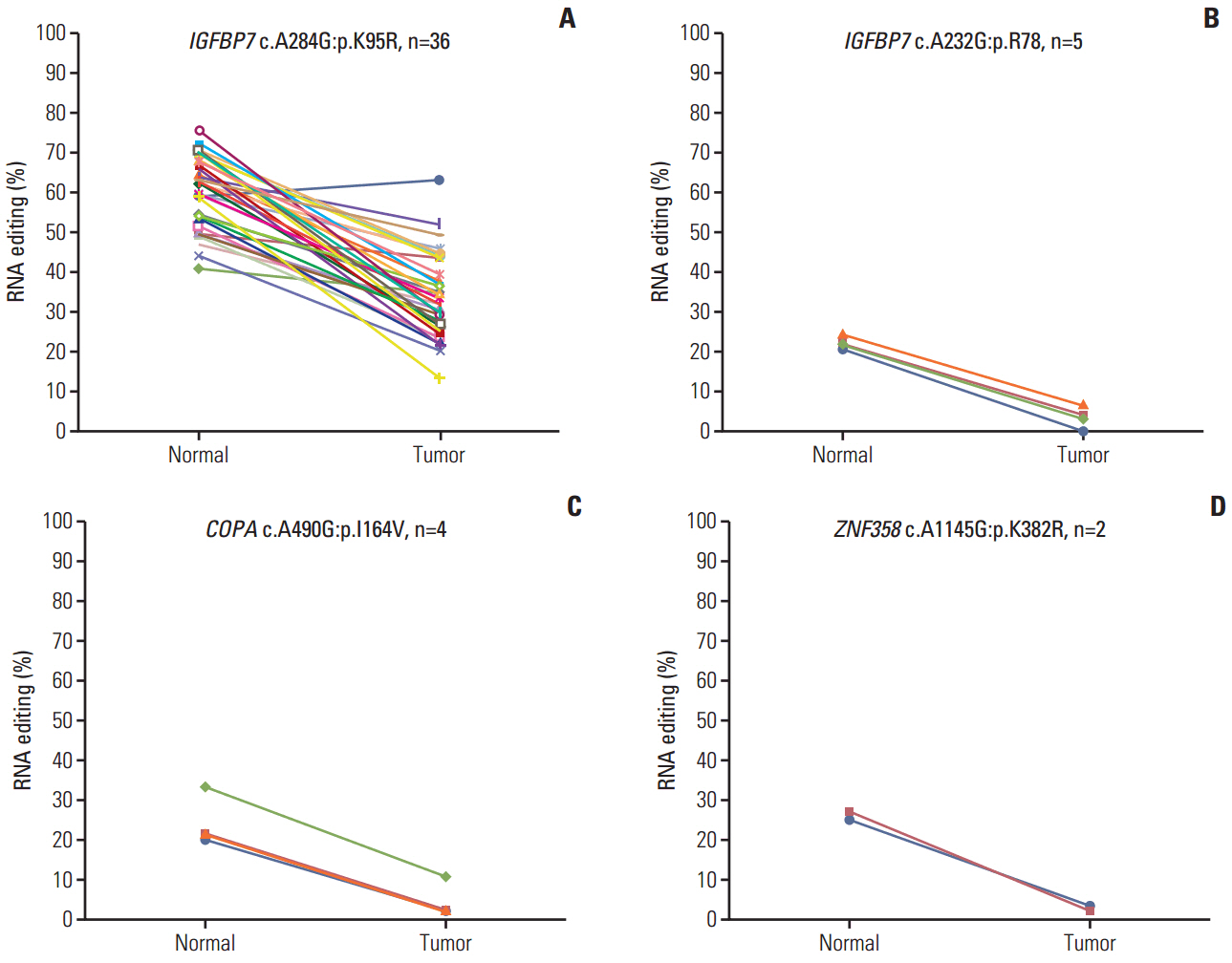
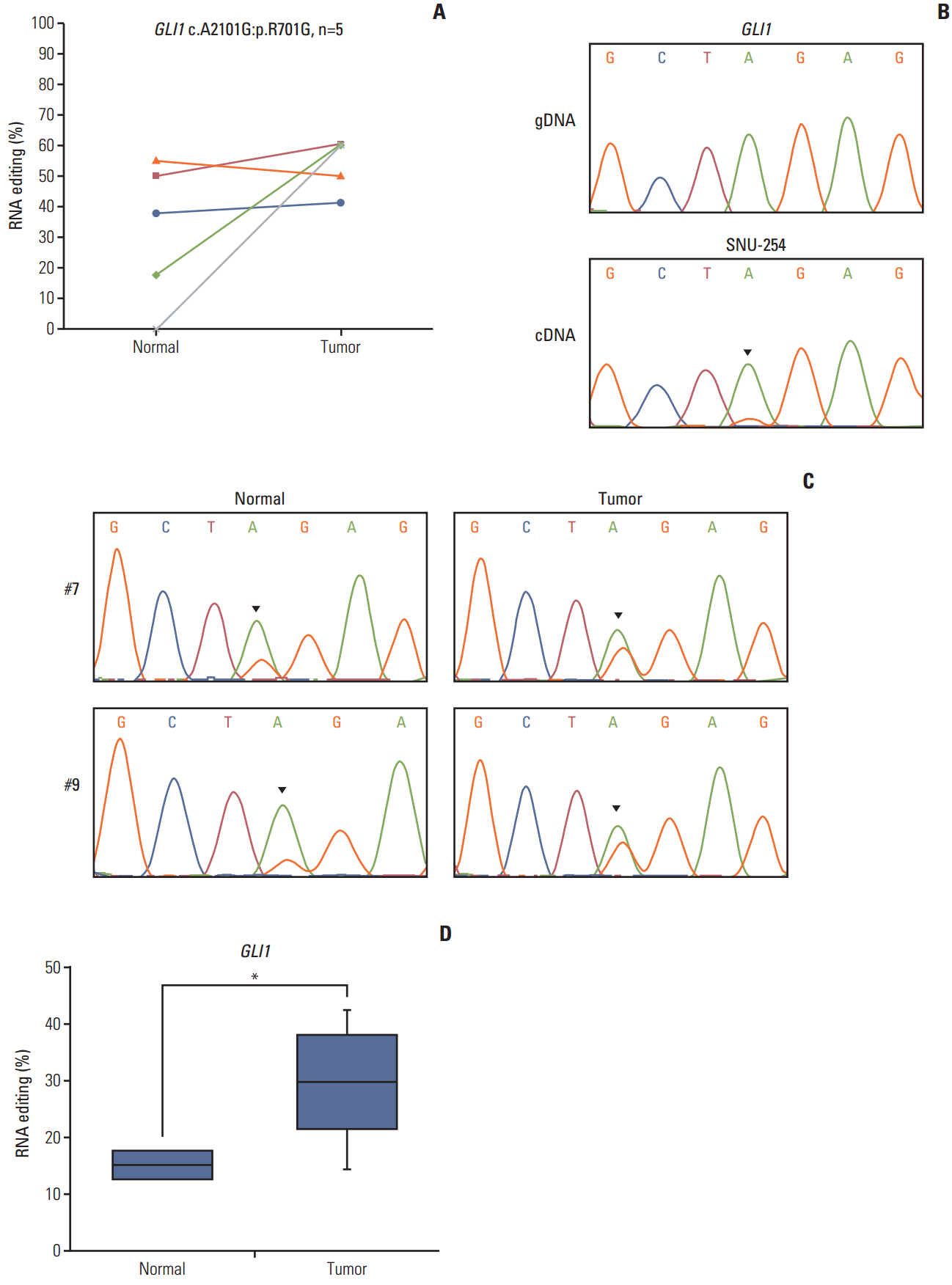
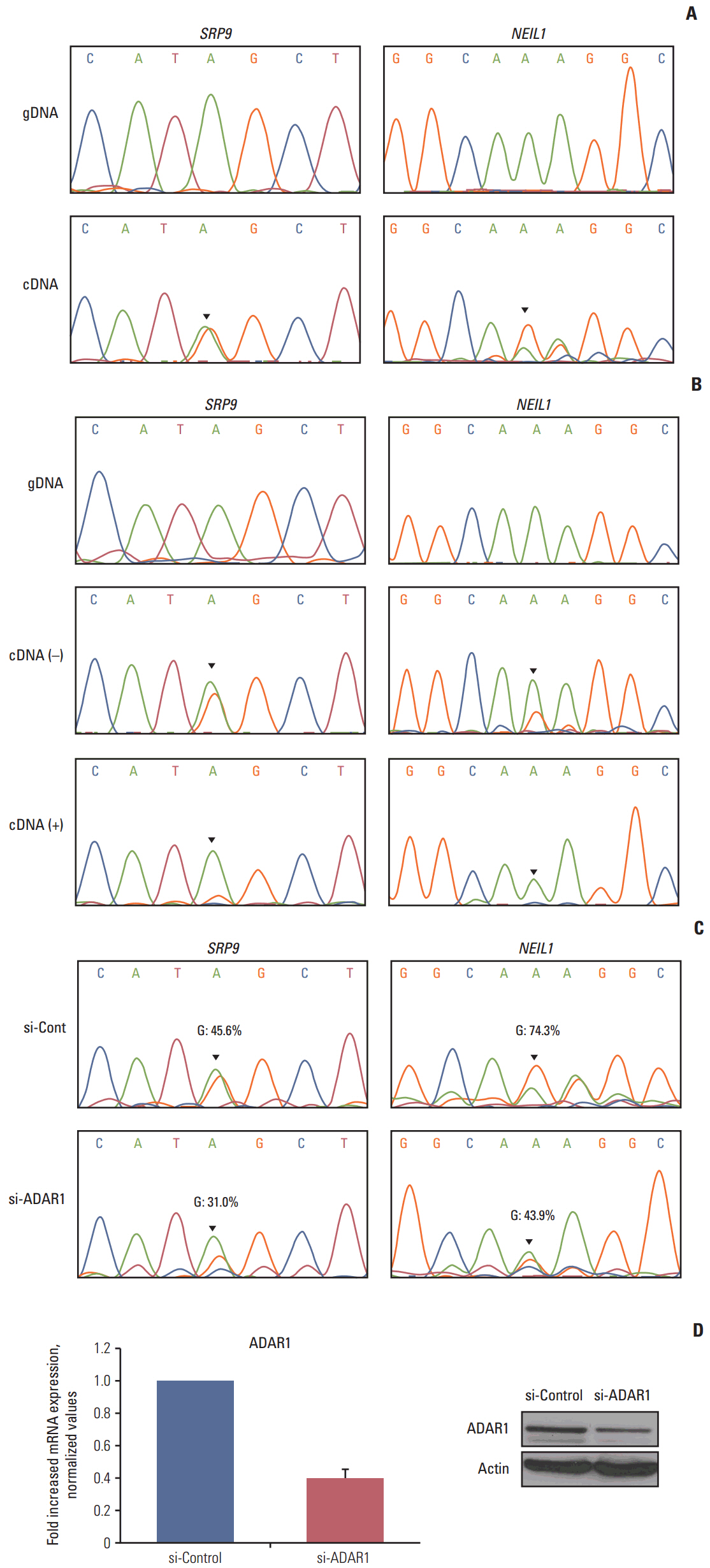
The number of RNA edits varied from 12 to 42 per sample. We also observed that hypoand hyper-RNA editing patterns were distinguishable within the samples. We found 10 recurrent nonsynonymous RNA editing candidates in nine genes (PDLIM, NEIL1, SRP9, GLI1, APMAP, IGFBP7, ZNF358, COPA, and ZNF587B) and validated some by Sanger sequencing and the inosine chemical erasing assay. We further showed that editing at these positions was performed by the adenosine deaminase acting on RNA 1 enzyme. Most of these genes are hypoedited in CRC, but editing of GLI1 was increased in cancer tissues compared with normal tissues.
CONCLUSION
Our results show that nonsynonymous RNA editing patterns can be used to identify CRC patients and could serve as novel biomarkers for CRC.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007; 446:153–8.
Article2. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.3. Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001; 2:869–78.
Article4. Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006; 7:919–31.
Article5. Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010; 79:321–49.
Article6. Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet. 2011; 43:745–52.
Article7. Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011; 333:53–8.
Article8. Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:D109–13.
Article9. Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010; 26:1772–6.
Article10. Kleinman CL, Adoue V, Majewski J. RNA editing of protein sequences: a rare event in human transcriptomes. RNA. 2012; 18:1586–96.
Article11. Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007; 17:1586–95.
Article12. Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008; 283:7251–60.13. Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001; 98:14687–92.
Article14. Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013; 19:209–16.
Article15. Han SW, Kim HP, Shin JY, Jeong EG, Lee WC, Kim KY, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med. 2014; 211:613–21.
Article16. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012; 22:568–76.
Article17. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.
Article18. Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat Chem Biol. 2010; 6:733–40.
Article19. Oh JH, Ku JL, Yoon KA, Kwon HJ, Kim WH, Park HS, et al. Establishment and characterization of 12 human colorectalcarcinoma cell lines. Int J Cancer. 1999; 81:902–10.
Article20. Han L, Diao L, Yu S, Xu X, Li J, Zhang R, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–28.
Article21. Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003; 278:1391–4.
Article22. Piskol R, Peng Z, Wang J, Li JB. Lack of evidence for existence of noncanonical RNA editing. Nat Biotechnol. 2013; 31:19–20.
Article23. Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A. 2010; 107:20715–9.
Article24. Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009; 461:809–13.
Article25. Verhagen HJ, de Leeuw DC, Roemer MG, Denkers F, Pouwels W, Rutten A, et al. IGFBP7 induces apoptosis of acute myeloid leukemia cells and synergizes with chemotherapy in suppression of leukemia cell survival. Cell Death Dis. 2014; 5:e1300.
Article26. Liu L, Yang Z, Zhang W, Yan B, Gu Q, Jiao J, et al. Decreased expression of IGFBP7 was a poor prognosis predictor for gastric cancer patients. Tumour Biol. 2014; 35:8875–81.
Article27. Chan TH, Lin CH, Qi L, Fei J, Li Y, Yong KJ, et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014; 63:832–43.
Article28. Shimokawa T, Rahman MF, Tostar U, Sonkoly E, Stahle M, Pivarcsi A, et al. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013; 10:321–33.
Article29. Bar EE, Chaudhry A, Farah MH, Eberhart CG. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am J Pathol. 2007; 170:347–55.30. Akiyoshi T, Nakamura M, Koga K, Nakashima H, Yao T, Tsuneyoshi M, et al. GLI1, downregulated in colorectal cancers, inhibits proliferation of colon cancer cells involving Wnt signalling activation. Gut. 2006; 55:991–9.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Bioinformatics Approaches for the Identification and Annotation of RNA Editing Sites
- Adenosine Deaminase in Human Skin
- Further Studies of Adenosine Deaminase in the Human Skin (II)
- Identification of neoantigens derived from alternative splicing and RNA modification
- Diagnostic Value of Adenosine Deaminase Activity in Tuberculous Pericardial Effusion