J Periodontal Implant Sci.
2016 Feb;46(1):2-9. 10.5051/jpis.2016.46.1.2.
Surface interactions between two of the main periodontal pathogens: Porphyromonas gingivalis and Tannerella forsythia
- Affiliations
-
- 1Formerly, Department of Medicine, University of California School of Medicine, Los Angeles, CA, USA.
- 2Departments of Dental Education and Periodontology, Dental Science Research Institute, Chonnam National University School of Dentistry, Gwangju, Korea. swlee@chonnam.ac.kr
- KMID: 2391725
- DOI: http://doi.org/10.5051/jpis.2016.46.1.2
Abstract
- PURPOSE
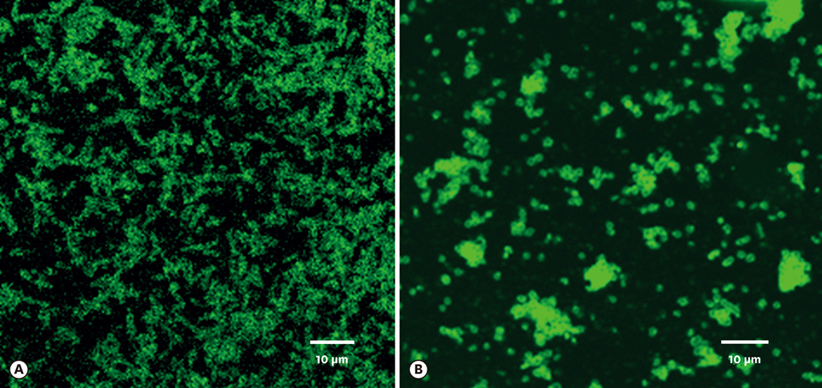
Porphyromonas gingivalis and Tannerella forsythia have been implicated as the major etiologic agents of periodontal disease. These two bacteria are frequently isolated together from the periodontal lesion, and it has been suggested that their interaction may increase each one's virulence potential. The purpose of this study was to identify proteins on the surface of these organisms that are involved in interbacterial binding.
METHODS
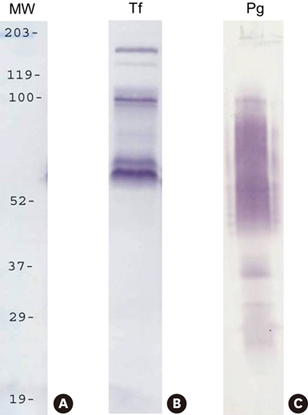
Biotin labeling of surface proteins of P. gingivalis and T. forsythia and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed to identify surface proteins involved in the coaggregating activity between P. gingivalis and T. forsythia.
RESULTS
It was found that three major T. forsythia proteins sized 161, 100, and 62 kDa were involved in binding to P. gingivalis, and P. gingivalis proteins sized 35, 32, and 26 kDa were involved in binding to T. forsythia cells.
CONCLUSIONS
LC-MS/MS analysis identified one T. forsythia surface protein (TonB-linked outer membrane protein) involved in interbacterial binding to P. gingivalis. However, the nature of other T. forsythia and P. gingivalis surface proteins identified by biotin labeling could not be determined. Further analysis of these proteins will help elucidate the molecular mechanisms that mediate coaggregation between P. gingivalis and T. forsythia.
Keyword
MeSH Terms
Figure
Reference
-
1. Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997; 61:136–169.
Article2. Marsh PD. Dental plaque: biological significance of a biofilm and community life-style. J Clin Periodontol. 2005; 32:Suppl 6. 7–15.
Article3. Marsh PD, Moter A, Devine DA. Dental plaque biofilms: communities, conflict and control. Periodontol 2000. 2011; 55:16–35.
Article4. Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001; 183:3770–3783.
Article5. Socransky SS, Haffajee AD. Evidence of bacterial etiology: a historical perspective. Periodontol 2000. 1994; 5:7–25.
Article6. Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci U S A. 1999; 96:14547–14552.
Article7. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005; 43:5721–5732.
Article8. Marsh PD. Dental plaque: biological significance of a biofilm and community life-style. J Clin Periodontol. 2005; 32:Suppl 6. 7–15.
Article9. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998; 25:134–144.
Article10. Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994; 5:78–111.
Article11. Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010; 8:481–490.
Article12. Kolenbrander PE. Coaggregations among oral bacteria. Methods Enzymol. 1995; 253:385–397.13. Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr. Communication among oral bacteria. Microbiol Mol Biol Rev. 2002; 66:486–505.
Article14. Kirschbaum M, Schultze-Mosgau S, Pfister W, Eick S. Mixture of periodontopathogenic bacteria influences interaction with KB cells. Anaerobe. 2010; 16:461–468.
Article15. Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000; 54:413–437.
Article16. Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, et al. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007; 75:1704–1712.
Article17. Zambon JJ. Periodontal diseases: microbial factors. Ann Periodontol. 1996; 1:879–925.
Article18. Kamma JJ, Nakou M, Manti FA. Predominant microflora of severe, moderate and minimal periodontal lesions in young adults with rapidly progressive periodontitis. J Periodontal Res. 1995; 30:66–72.
Article19. Yoneda M, Hirofuji T, Anan H, Matsumoto A, Hamachi T, Nakayama K, et al. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J Periodontal Res. 2001; 36:237–243.
Article20. Takemoto T, Kurihara H, Dahlen G. Characterization of Bacteroides forsythus isolates. J Clin Microbiol. 1997; 35:1378–1381.21. Yoneda M, Yoshikane T, Motooka N, Yamada K, Hisama K, Naito T, et al. Stimulation of growth of Porphyromonas gingivalis by cell extracts from Tannerella forsythia . J Periodontal Res. 2005; 40:105–109.
Article22. Yao ES, Lamont RJ, Leu SP, Weinberg A. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol. 1996; 11:35–41.
Article23. Um HS, Lee SW, Park JH, Nauman RK. Coaggregation between Porphyromonas gingivalis and Tannerella forsythia . J Korean Acad Periodontol. 2006; 36:265–272.
Article24. Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003; 4:397–407.
Article25. Kamaguchi A, Ohyama T, Sakai E, Nakamura R, Watanabe T, Baba H, et al. Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia . Microbiology. 2003; 149:1257–1264.
Article26. Shimotahira N, Oogai Y, Kawada-Matsuo M, Yamada S, Fukutsuji K, Nagano K, et al. The surface layer of Tannerella forsythia contributes to serum resistance and oral bacterial coaggregation. Infect Immun. 2013; 81:1198–1206.
Article27. Sabet M, Lee SW, Nauman RK, Sims T, Um HS. The surface (S-) layer is a virulence factor of Bacteroides forsythus . Microbiology. 2003; 149:3617–3627.28. Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia . Gene. 2006; 371:102–111.
Article29. Yoo JY, Kim HC, Zhu W, Kim SM, Sabet M, Handfield M, et al. Identification of Tannerella forsythia antigens specifically expressed in patients with periodontal disease. FEMS Microbiol Lett. 2007; 275:344–352.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Coaggregation between Porphyromonas gingivalis and Tannerella forsythia
- Visualization of periodontopathic bacteria within crevicular epithelial cells with fluorescence in situ hybridization
- Effects of Microbial Communication on The Growth of Periodontopathogens
- Subgingival pathogens in chronic periodontitis patients affected by type 2 diabetes mellitus: a retrospective case-control study
- Inhibition of biofilm formation of periodontal pathogens by D-Arabinose