J Breast Cancer.
2017 Jun;20(2):198-202. 10.4048/jbc.2017.20.2.198.
Epigenetic Silencing of MORT Is an Early Event in Cancer and Is Associated with Luminal, Receptor Positive Breast Tumor Subtypes
- Affiliations
-
- 1The University of Arizona Cancer Center, Tucson, USA. lvrba@uacc.arizona.edu
- 2Department of Pharmacology & Toxicology, College of Pharmacy, The University of Arizona, Tucson, USA.
- KMID: 2389758
- DOI: http://doi.org/10.4048/jbc.2017.20.2.198
Abstract
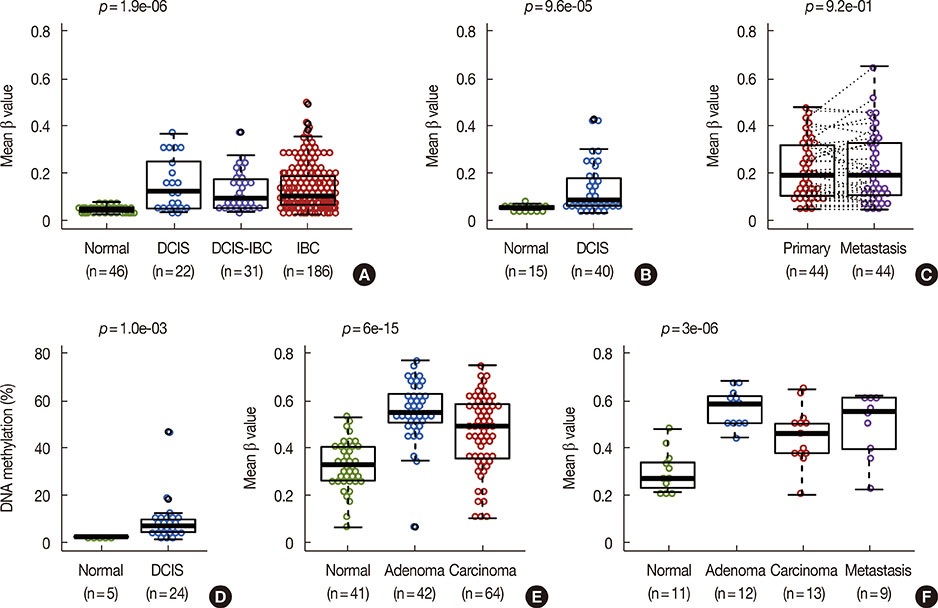
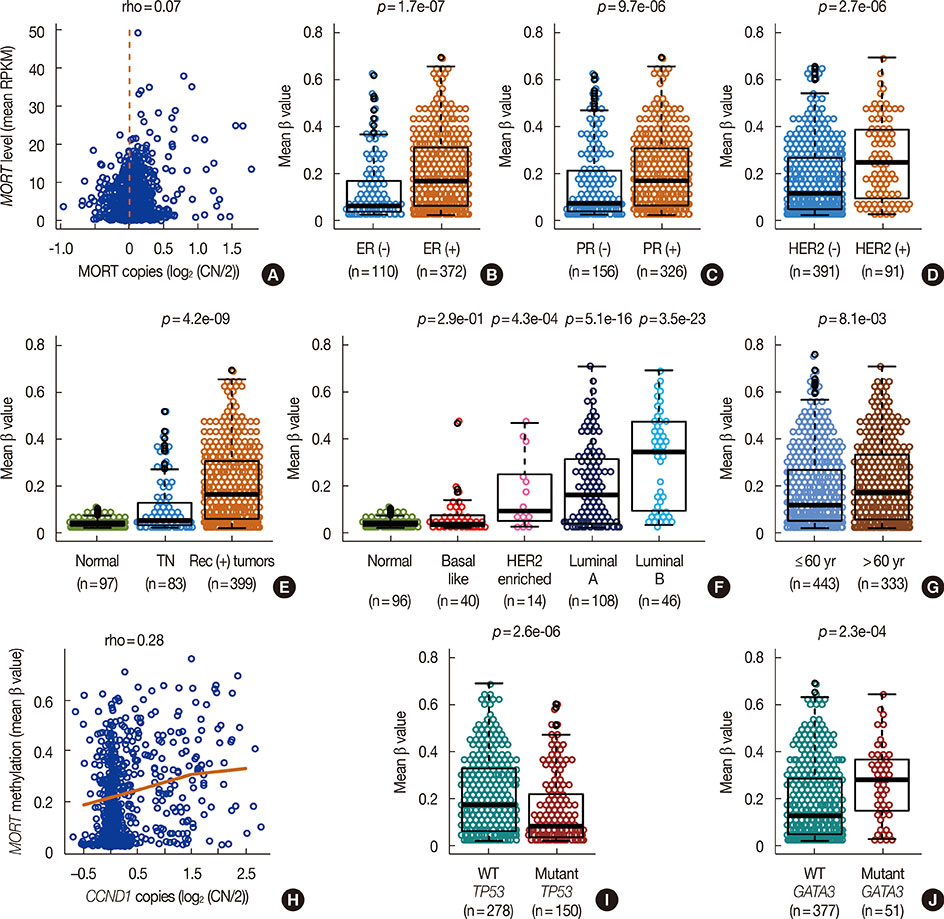
- Immortality is an essential characteristic of cancer cells; a recent transcriptomic study of epithelial cell immortalization has linked epigenetic silencing of the long noncoding RNA Mortal Obligate RNA Transcript (MORT; alias ZNF667-AS1) to this process. This study evaluated the epigenetic and transcriptional state of MORT in two premalignant conditions"”ductal carcinomas in situ and colon adenomas. Results show that MORT silencing is an early epigenetic event in human carcinogenesis, likely occurring near the point where premalignant cells gain immortality; this epigenetic silencing is maintained throughout malignant transformation and metastatic growth. Additional associations between MORT loss and clinical and molecular features of breast tumors showed that silencing of MORT occurs predominantly in luminal, receptor-positive breast cancer; is associated with overexpression of CCND1 and mutations of GATA3; and is negatively correlated with TP53 mutations. Taken in toto, MORT silencing occurs early in breast carcinogenesis, probably during cellular immortalization, and precedes the development of invasive luminal breast cancer.
MeSH Terms
Figure
Reference
-
1. Vrba L, Garbe JC, Stampfer MR, Futscher BW. A lincRNA connected to cell mortality and epigenetically-silenced in most common human cancers. Epigenetics. 2015; 10:1074–1083.
Article2. Allred DC, Mohsin SK. Biological features of premalignant disease in the human breast. J Mammary Gland Biol Neoplasia. 2000; 5:351–364.3. Yashima K, Milchgrub S, Gollahon LS, Maitra A, Saboorian MH, Shay JW, et al. Telomerase enzyme activity and RNA expression during the multistage pathogenesis of breast carcinoma. Clin Cancer Res. 1998; 4:229–234.4. Shpitz B, Zimlichman S, Zemer R, Bomstein Y, Zehavi T, Liverant S, et al. Telomerase activity in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 1999; 58:65–69.
Article5. Umbricht CB, Sherman ME, Dome J, Carey LA, Marks J, Kim N, et al. Telomerase activity in ductal carcinoma in situ and invasive breast cancer. Oncogene. 1999; 18:3407–3414.
Article6. R: a language and environment for statistical computing. R Foundation for Statistical Computing. 2017. Accessed April 27th. http://www.R-project.org/.7. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995; 57:289–300.
Article8. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.9. Fleischer T, Frigessi A, Johnson KC, Edvardsen H, Touleimat N, Klajic J, et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014; 15:435.
Article10. Johnson KC, Koestler DC, Fleischer T, Chen P, Jenson EG, Marotti JD, et al. DNA methylation in ductal carcinoma in situ related with future development of invasive breast cancer. Clin Epigenetics. 2015; 7:75.
Article11. Reyngold M, Turcan S, Giri D, Kannan K, Walsh LA, Viale A, et al. Remodeling of the methylation landscape in breast cancer metastasis. PLoS One. 2014; 9:e103896.
Article12. Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014; 147:418–429.
Article13. Qu X, Sandmann T, Frierson H Jr, Fu L, Fuentes E, Walter K, et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene. 2016; 35:6403–6415.
Article14. Abba MC, Gong T, Lu Y, Lee J, Zhong Y, Lacunza E, et al. A molecular portrait of high-grade ductal carcinoma in situ. Cancer Res. 2015; 75:3980–3990.
Article15. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009; 27:1160–1167.
Article16. Jenkins EO, Deal AM, Anders CK, Prat A, Perou CM, Carey LA, et al. Age-specific changes in intrinsic breast cancer subtypes: a focus on older women. Oncologist. 2014; 19:1076–1083.
Article17. Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008; 8:714–724.
Article18. Garbe JC, Vrba L, Sputova K, Fuchs L, Novak P, Brothman AR, et al. Immortalization of normal human mammary epithelial cells in two steps by direct targeting of senescence barriers does not require gross genomic alterations. Cell Cycle. 2014; 13:3423–3435.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Subtype Is a Predictive Factor of Nonsentinel Lymph Node Involvement in Sentinel Node-Positive Breast Cancer Patients
- Potential Benefits of Neoadjuvant Chemotherapy in Clinically Node-Positive Luminal Subtypeâ» Breast Cancer
- Cancer Subtypes of Breast Carcinoma with Micropapillary and Mucinous Component Based on Immunohistochemical Profile
- Prognosis according to the timing of recurrence in breast cancer
- Genomic Profiling Shows Increased Glucose Metabolism in Luminal B Breast Cancer