Obstet Gynecol Sci.
2017 May;60(3):241-259. 10.5468/ogs.2017.60.3.241.
Autophagy in the placenta
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. crroh@skku.edu
- KMID: 2378586
- DOI: http://doi.org/10.5468/ogs.2017.60.3.241
Abstract
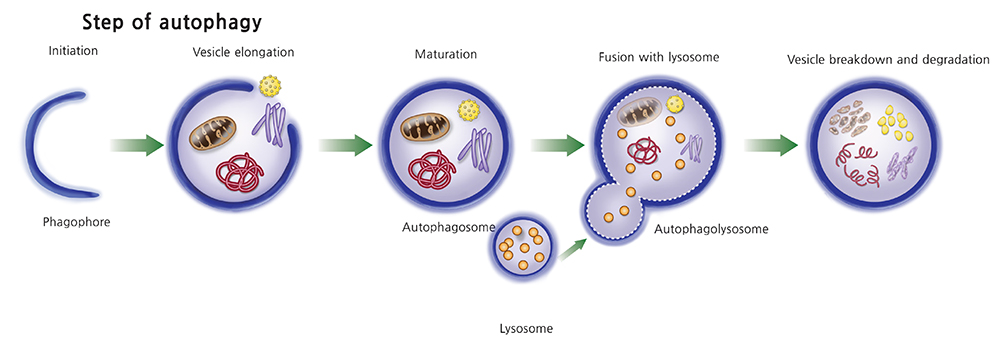
- Autophagy is an evolutionarily conserved catalytic process by which cytoplasmic components including damaged macromolecules and organelles are degraded. The role of autophagy includes adaptive responses to nutrition deprivation or intracellular stimuli. Although autophagosomes were first observed in early 1960s, it was 1990s that autophagy-related genes in yeast were identified and studied. Nowadays, the molecular machinery of autophagy and signaling pathway to various stimuli are almost outlined. Dysregulation of autophagic activity has been implicated in many human diseases including neurodegenerative diseases, infection and inflammation, and malignancies. However, since current understanding of autophagy in placenta is just at the beginning, this paper aims to provide general information on autophagy (part I) and to summarize articles on autophagy in human placenta (part II). This review article will serve as a basis for further researches on autophagy in relation to human pregnancy and its complications.
Keyword
MeSH Terms
Figure
Reference
-
1. Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004; 23:2891–2906.2. Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008; 9:1004–1010.3. Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011; 7:279–296.4. Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962; 12:198–202.5. Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014; 24:9–23.6. Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992; 119:301–311.7. Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014; 24:24–41.8. Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007; 39:596–604.9. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999; 402:672–676.10. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013; 368:1845–1846.11. Ma JF, Huang Y, Chen SD, Halliday G. Immunohistochemical evidence for macroautophagy in neurones and endothelial cells in Alzheimer's disease. Neuropathol Appl Neurobiol. 2010; 36:312–319.12. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011; 469:323–335.13. Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008; 321:117–120.14. Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004; 432:1032–1036.15. Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010; 40:280–293.16. Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010; 22:124–131.17. He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010; 22:140–149.18. Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001; 2:211–216.19. Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010; 22:157–168.20. Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, et al. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 2010; 298:E751–E760.21. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010; 120:127–141.22. Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009; 69:4415–4423.23. Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007; 27:135–144.24. Harris J. Autophagy and cytokines. Cytokine. 2011; 56:140–144.25. Jia G, Cheng G, Gangahar DM, Agrawal DK. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol. 2006; 84:448–454.26. Sivaprasad U, Basu A. Inhibition of ERK attenuates autophagy and potentiates tumour necrosis factor-alpha-induced cell death in MCF-7 cells. J Cell Mol Med. 2008; 12:1265–1271.27. Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009; 29:2570–2581.28. DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008; 22:239–251.29. Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010; 22:177–180.30. Liu L, Wise DR, Diehl JA, Simon MC. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem. 2008; 283:31153–31162.31. Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007; 26:1749–1760.32. Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013; 27:1016–1031.33. Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009; 458:1127–1130.34. Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004; 36:2491–2502.35. Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004; 15:1101–1111.36. Esteban-Martinez L, Boya P. Autophagic flux determination in vivo and ex vivo. Methods. 2015; 75:79–86.37. Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. J Pathol. 2010; 221:117–124.38. Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007; 3:452–460.39. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000; 19:5720–5728.40. Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004; 117(Pt 13):2805–2812.41. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012; 8:445–544.42. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010; 140:313–326.43. Jiang P, Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015; 75:13–18.44. Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007; 3:542–545.45. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007; 282:24131–24145.46. Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009; 137:1001–1004.47. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007; 8:741–752.48. Ravikumar B, Berger Z, Vacher C, O'Kane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006; 15:1209–1216.49. Jiang J, Jiang J, Zuo Y, Gu Z. Rapamycin protects the mitochondria against oxidative stress and apoptosis in a rat model of Parkinson's disease. Int J Mol Med. 2013; 31:825–832.50. Cai N, Zhao X, Jing Y, Sun K, Jiao S, Chen X, et al. Autophagy protects against palmitate-induced apoptosis in hepatocytes. Cell Biosci. 2014; 4:28.51. Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014; 15:81–94.52. Amir M, Zhao E, Fontana L, Rosenberg H, Tanaka K, Gao G, et al. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 2013; 20:878–887.53. Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010; 17:268–277.54. Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006; 116:2161–2172.55. Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012; 287:12455–12468.56. Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013; 140:1321–1329.57. Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK. Autophagy and apoptosis: where do they meet? Apoptosis. 2014; 19:555–566.58. Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006; 126:121–134.59. Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009; 137:721–735.60. Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009; 11:1355–1362.61. Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006; 8:1124–1132.62. Morris HR. Genetics of Parkinson's disease. Ann Med. 2005; 37:86–96.63. Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012; 135(Pt 7):2169–2177.64. Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, et al. α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010; 190:1023–1037.65. Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007; 450:1253–1257.66. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010; 32:227–239.67. Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012; 150:803–815.68. Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009; 15:267–276.69. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008; 40:955–962.70. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003; 112:1809–1820.71. Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011; 25:717–729.72. Chen S, Rehman SK, Zhang W, Wen A, Yao L, Zhang J. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010; 1806:220–229.73. Kim KH, Lee MS. Autophagy: a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014; 10:322–337.74. Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008; 8:325–332.75. Quan W, Hur KY, Lim Y, Oh SH, Lee JC, Kim KH, et al. Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2012; 55:392–403.76. Tomic T, Botton T, Cerezo M, Robert G, Luciano F, Puissant A, et al. Metformin inhibits melanoma development through autophagy and apoptosis mechanisms. Cell Death Dis. 2011; 2:e199.77. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009; 458:1131–1135.78. Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013; 20:3–11.79. Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003; 100:15077–15082.80. Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007; 128:931–946.81. Wada Y, Sun-Wada GH, Kawamura N. Microautophagy in the visceral endoderm is essential for mouse early development. Autophagy. 2013; 9:252–254.82. Sato M, Sato K. Dynamic regulation of autophagy and endocytosis for cell remodeling during early development. Traffic. 2013; 14:479–486.83. Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011; 334:1141–1144.84. Sato M, Sato K. Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy. 2012; 8:424–425.85. Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007; 316:407–408.86. Song BS, Yoon SB, Kim JS, Sim BW, Kim YH, Cha JJ, et al. Induction of autophagy promotes preattachment development of bovine embryos by reducing endoplasmic reticulum stress. Biol Reprod. 2012; 87:81–11.87. Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007; 447:679–685.88. Weitlauf HM. Metabolic changes in the blastocysts of mice and rats during delayed implantation. J Reprod Fertil. 1974; 39:213–224.89. Lee JE, Oh HA, Song H, Jun JH, Roh CR, Xie H, et al. Autophagy regulates embryonic survival during delayed implantation. Endocrinology. 2011; 152:2067–2075.90. Kamei T, Jones SR, Chapman BM, MCGonigle KL, Dai G, Soares MJ. The phosphatidylinositol 3-kinase/Akt signaling pathway modulates the endocrine differentiation of trophoblast cells. Mol Endocrinol. 2002; 16:1469–1481.91. Avagliano L, Terraneo L, Virgili E, Martinelli C, Doi P, Samaja M, et al. Autophagy in normal and abnormal early human pregnancies. Reprod Sci. 2015; 22:838–844.92. Hung TH, Hsieh TT, Chen SF, Li MJ, Yeh YL. Autophagy in the human placenta throughout gestation. PLoS One. 2013; 8:e83475.93. Oh SY, Choi SJ, Kim KH, Cho EY, Kim JH, Roh CR. Autophagy-related proteins, LC3 and Beclin-1, in placentas from pregnancies complicated by preeclampsia. Reprod Sci. 2008; 15:912–920.94. Akaishi R, Yamada T, Nakabayashi K, Nishihara H, Furuta I, Kojima T, et al. Autophagy in the placenta of women with hypertensive disorders in pregnancy. Placenta. 2014; 35:974–980.95. Hung TH, Chen SF, Lo LM, Li MJ, Yeh YL, Hsieh TT. Increased autophagy in placentas of intrauterine growth-restricted pregnancies. PLoS One. 2012; 7:e40957.96. Curtis S, Jones CJ, Garrod A, Hulme CH, Heazell AE. Identification of autophagic vacuoles and regulators of autophagy in villous trophoblast from normal term pregnancies and in fetal growth restriction. J Matern Fetal Neonatal Med. 2013; 26:339–346.97. Gao L, Qi HB, Kamana KC, Zhang XM, Zhang H, Baker PN. Excessive autophagy induces the failure of trophoblast invasion and vasculature: possible relevance to the pathogenesis of preeclampsia. J Hypertens. 2015; 33:106–117.98. Goldman-Wohl D, Cesla T, Smith Y, Greenfield C, Dechend R, Staff AC, et al. Expression profiling of autophagy associated genes in placentas of preeclampsia. Placenta. 2013; 34:959–962.99. Burton GJ, Yung HW, Murray AJ. Mitochondrial - Endoplasmic reticulum interactions in the trophoblast: Stress and senescence. Placenta. 2016; 04. 04. [Epub]. DOI: 10.1016/j.placenta.2016.04.001.100. Kanninen TT, Jayaram A, Jaffe Lifshitz S, Witkin SS. Altered autophagy induction by sera from pregnant women with pre-eclampsia: a case-control study. BJOG. 2014; 121:958–964.101. Cheng SB, Nakashima A, Sharma S. Understanding pre-eclampsia using Alzheimer's etiology: an intriguing viewpoint. Am J Reprod Immunol. 2016; 75:372–381.102. Watada H, Fujitani Y. Minireview: autophagy in pancreatic β-cells and its implication in diabetes. Mol Endocrinol. 2015; 29:338–348.103. Chen B, Longtine MS, Nelson DM. Hypoxia induces autophagy in primary human trophoblasts. Endocrinology. 2012; 153:4946–4954.104. Nakashima A, Yamanaka-Tatematsu M, Fujita N, Koizumi K, Shima T, Yoshida T, et al. Impaired autophagy by soluble endoglin, under physiological hypoxia in early pregnant period, is involved in poor placentation in preeclampsia. Autophagy. 2013; 9:303–316.105. Yamanaka-Tatematsu M, Nakashima A, Fujita N, Shima T, Yoshimori T, Saito S. Autophagy induced by HIF1α overexpression supports trophoblast invasion by supplying cellular energy. PLoS One. 2013; 8:e76605.106. Melland-Smith M, Ermini L, Chauvin S, Craig-Barnes H, Tagliaferro A, Todros T, et al. Disruption of sphingolipid metabolism augments ceramide-induced autophagy in preeclampsia. Autophagy. 2015; 11:653–669.107. Crocker I. Gabor Than Award Lecture 2006: pre-eclampsia and villous trophoblast turnover. Perspectives and possibilities. Placenta. 2007; 28:Suppl A. S4–S13.108. Choi JH, Lee HJ, Yang TH, Kim GJ. Effects of hypoxia inducible factors-1α on autophagy and invasion of trophoblasts. Clin Exp Reprod Med. 2012; 39:73–80.109. Jansson T, Aye IL, Goberdhan DC. The emerging role of mTORC1 signaling in placental nutrient-sensing. Placenta. 2012; 33:Suppl 2. e23–e29.110. Roos S, Jansson N, Palmberg I, Saljo K, Powell TL, Jansson T. Mammalian target of rapamycin in the human placenta regulates leucine transport and is down-regulated in restricted fetal growth. J Physiol. 2007; 582(Pt 1):449–459.111. Pollheimer J, Knofler M. Signalling pathways regulating the invasive differentiation of human trophoblasts: a review. Placenta. 2005; 26:Suppl A. S21–S30.112. Ito S, Koshikawa N, Mochizuki S, Takenaga K. 3-Methyladenine suppresses cell migration and invasion of HT1080 fibrosarcoma cells through inhibiting phosphoinositide 3-kinases independently of autophagy inhibition. Int J Oncol. 2007; 31:261–268.113. Milano V, Piao Y, LaFortune T, de Groot J. Dasatinib-induced autophagy is enhanced in combination with temozolomide in glioma. Mol Cancer Ther. 2009; 8:394–406.114. Indelicato M, Pucci B, Schito L, Reali V, Aventaggiato M, Mazzarino MC, et al. Role of hypoxia and autophagy in MDA-MB-231 invasiveness. J Cell Physiol. 2010; 223:359–368.115. Wang Y, Wang C, Tang H, Wang M, Weng J, Liu X, et al. Decrease of autophagy activity promotes malignant progression of tongue squamous cell carcinoma. J Oral Pathol Med. 2013; 42:557–564.116. Nakabayashi Y, Nakashima A, Yoshino O, Shima T, Shiozaki A, Adachi T, et al. Impairment of the accumulation of decidual T cells, NK cells, and monocytes, and the poor vascular remodeling of spiral arteries, were observed in oocyte donation cases, regardless of the presence or absence of preeclampsia. J Reprod Immunol. 2016; 114:65–74.117. Crocker IP, Cooper S, Ong SC, Baker PN. Differences in apoptotic susceptibility of cytotrophoblasts and syncytiotrophoblasts in normal pregnancy to those complicated with preeclampsia and intrauterine growth restriction. Am J Pathol. 2003; 162:637–643.118. Huppertz B, Kadyrov M, Kingdom JC. Apoptosis and its role in the trophoblast. Am J Obstet Gynecol. 2006; 195:29–39.119. Ishihara N, Matsuo H, Murakoshi H, Laoag-Fernandez JB, Samoto T, Maruo T. Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardation. Am J Obstet Gynecol. 2002; 186:158–166.120. Cha HH, Hwang JR, Kim HY, Choi SJ, Oh SY, Roh CR. Autophagy induced by tumor necrosis factor α mediates intrinsic apoptosis in trophoblastic cells. Reprod Sci. 2014; 21:612–622.121. Signorelli P, Avagliano L, Virgili E, Gagliostro V, Doi P, Braidotti P, et al. Autophagy in term normal human placentas. Placenta. 2011; 32:482–485.122. Doulaveris G, Orfanelli T, Benn K, Zervoudakis I, Skupski D, Witkin SS. A polymorphism in an autophagy-related gene, ATG16L1, influences time to delivery in women with an unfavorable cervix who require labor induction. J Perinat Med. 2013; 41:411–414.123. Avagliano L, Virgili E, Garo C, Quadrelli F, Doi P, Samaja M, et al. Autophagy and human parturition: evaluation of LC3 expression in placenta from spontaneous or medically induced onset of labor. Biomed Res Int. 2013; 2013:689768.124. Brickle A, Tran HT, Lim R, Liong S, Lappas M. Autophagy, which is decreased in labouring fetal membranes, regulates IL-1β production via the inflammasome. Placenta. 2015; 36:1393–1404.125. Avagliano L, Massa V, Zullino S, Doi P, Marconi AM, Ferrazzi E, et al. Inflammation modulates LC3 expression in human preterm delivery. J Matern Fetal Neonatal Med. 2017; 30:698–704.126. Hirota Y, Cha J, Yoshie M, Daikoku T, Dey SK. Heightened uterine mammalian target of rapamycin complex 1 (mTORC1) signaling provokes preterm birth in mice. Proc Natl Acad Sci U S A. 2011; 108:18073–18078.127. Agrawal V, Jaiswal MK, Mallers T, Katara GK, Gilman-Sachs A, Beaman KD, et al. Altered autophagic flux enhances inflammatory responses during inflammation-induced preterm labor. Sci Rep. 2015; 5:9410.128. de Andrade Ramos BR, Witkin SS. The influence of oxidative stress and autophagy cross regulation on pregnancy outcome. Cell Stress Chaperones. 2016; 21:755–762.129. Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013; 110:12048–12053.130. Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009; 5:816–823.131. Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014; 10:70–79.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The role of autophagy in the placenta as a regulator of cell death
- The role of autophagy in cell proliferation and differentiation during tooth development
- Effects of hypoxia inducible factors-1alpha on autophagy and invasion of trophoblasts
- A Case of Placenta Percreta Involving the Urinary Bladder
- Neuronal Autophagy: Characteristic Features and Roles in Neuronal Pathophysiology