Cancer Res Treat.
2017 Apr;49(2):313-321. 10.4143/crt.2016.098.
Expression of Myxovirus Resistance A (MxA) Is Associated with Tumor-Infiltrating Lymphocytes in Human Epidermal Growth Factor Receptor 2 (HER2)–Positive Breast Cancers
- Affiliations
-
- 1Department of Pathology, BioMedical Research Institute, Pusan National University Hospital, Busan, Korea. ahrong2h@naver.com
- 2Department of Pathology, Pusan National University Yangsan Hospital, Yangsan, Korea. artinus2000@naver.com
- 3Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2378103
- DOI: http://doi.org/10.4143/crt.2016.098
Abstract
- PURPOSE
The prognostic significance of tumor-infiltrating lymphocytes (TILs) has been determined in breast cancers. Interferons can affect T-cell activity through direct and indirect mechanisms. Myxovirus resistance A (MxA) is an excellent marker of interferon activity. Here,we evaluated TILs and MxA expression in human epidermal growth factor receptor 2 (HER2)-positive breast cancers.
MATERIALS AND METHODS
Ninety cases of hormone receptor (HR)+/HER2+ tumors and 78 cases of HR-/HER2+ tumors were included. The TILs level was assessed using hematoxylin and eosin-stained full face sections, and MxA expressionwas evaluated by immunohistochemistrywith a tissue microarray.
RESULTS
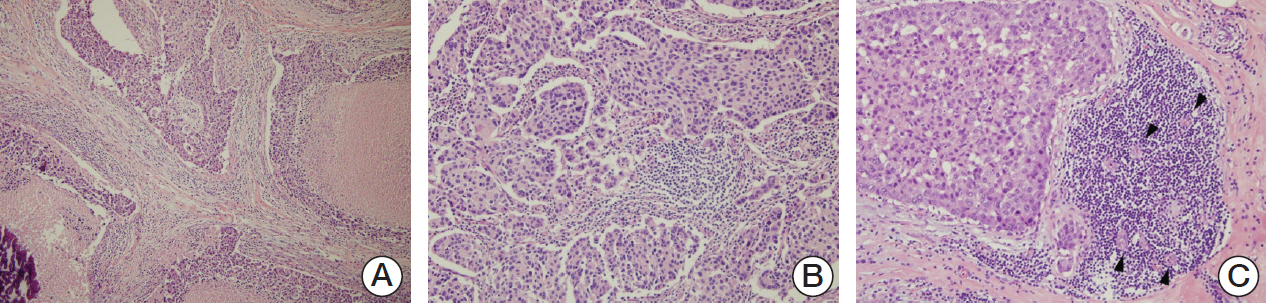
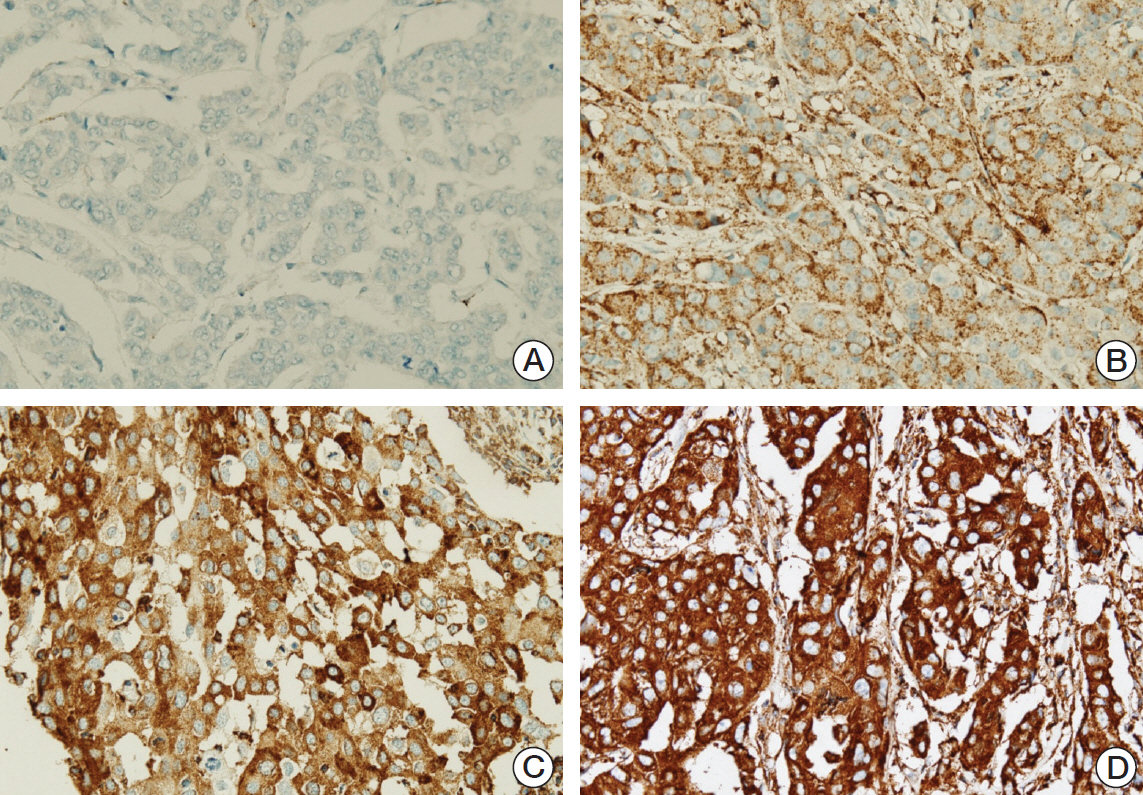
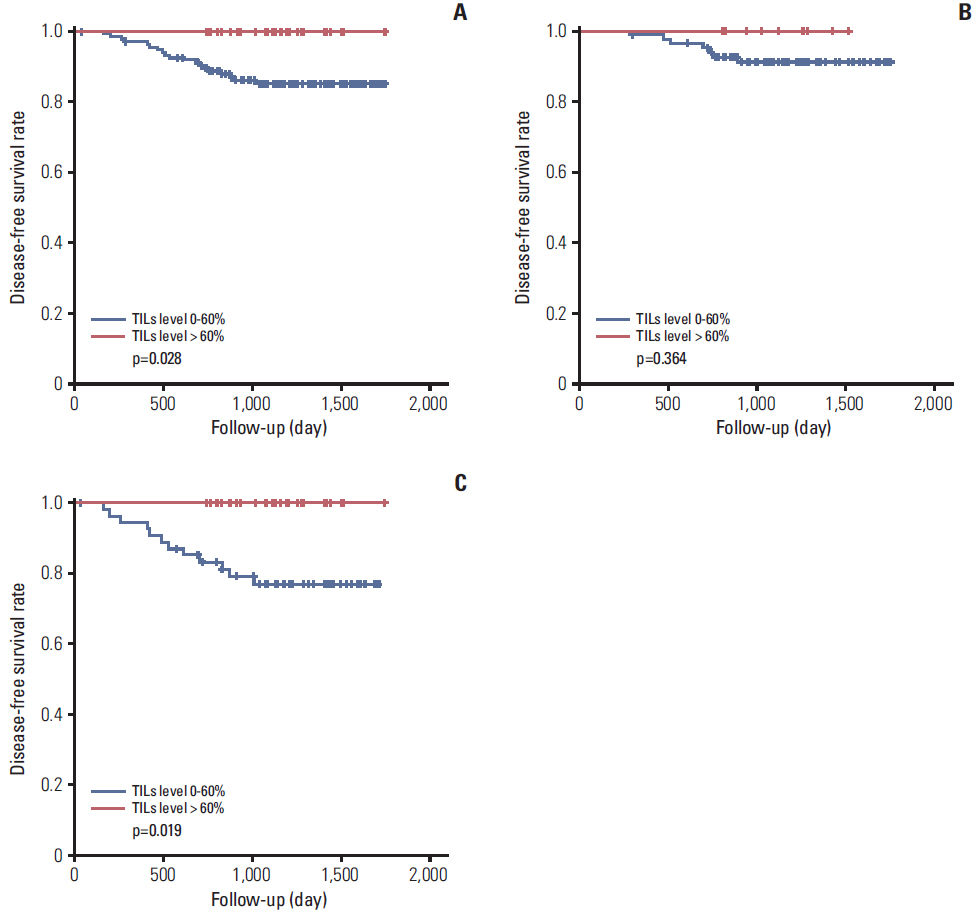
MxA protein expression was significantly higher in tumors with high histologic grade (p=0.023) and high levels of TILs (p=0.002). High levels of TILs were correlated with high histological grade (p=0.001), negative lymphovascular invasion (p=0.007), negative lymph node metastasis (p=0.007), absence of HR expression (p < 0.001), abundant tertiary lymphoid structures (TLSs) around ductal carcinoma in situ (p=0.018), and abundant TLSs around the invasive component (p < 0.001). High levels of TILs were also associated with improved disease-free survival, particularly in HR-/HER2+ breast cancers. However, MxA was not a prognostic factor.
CONCLUSION
High expression of MxA in tumor cells was associated with high levels of TILs in HER2-positive breast cancers. Additionally, a high level of TILs was a prognostic factor for breast cancer, whereas the level of MxA expression had no prognostic value.
MeSH Terms
-
Breast Neoplasms
Breast*
Carcinoma, Intraductal, Noninfiltrating
Disease-Free Survival
Epidermal Growth Factor*
Hematoxylin
Humans*
Interferons
Lymph Nodes
Lymphocytes, Tumor-Infiltrating*
Myxovirus Resistance Proteins
Neoplasm Metastasis
Orthomyxoviridae*
Receptor, Epidermal Growth Factor*
T-Lymphocytes
Epidermal Growth Factor
Hematoxylin
Interferons
Receptor, Epidermal Growth Factor
Figure
Cited by 1 articles
-
Expression of Immunoproteasome Subunit LMP7 in Breast Cancer and Its Association with Immune-Related Markers
Miseon Lee, In Hye Song, Sun-Hee Heo, Young-Ae Kim, In Ah Park, Won Seon Bang, Hye Seon Park, Gyungyub Gong, Hee Jin Lee
Cancer Res Treat. 2019;51(1):80-89. doi: 10.4143/crt.2017.500.
Reference
-
References
1. Ahn SG, Jeong J, Hong S, Jung WH. Current issues and clinical evidence in tumor-infiltrating lymphocytes in breast cancer. J Pathol Transl Med. 2015; 49:355–63.
Article2. Stagg J, Loi S, Divisekera U, Ngiow SF, Duret H, Yagita H, et al. Anti-ErbB-2 mAb therapy requires type I and II interferons and synergizes with anti-PD-1 or anti-CD137 mAb therapy. Proc Natl Acad Sci U S A. 2011; 108:7142–7.
Article3. Lee HJ, Kim JY, Park IA, Song IH, Yu JH, Ahn JH, et al. Prognostic significance of tumor-infiltrating lymphocytes and the tertiary lymphoid structures in HER2-positive breast cancer treated with adjuvant trastuzumab. Am J Clin Pathol. 2015; 144:278–88.
Article4. Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015; 26:259–71.
Article5. Schlee M. Master sensors of pathogenic RNA: RIG-I like receptors. Immunobiology. 2013; 218:1322–35.6. Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008; 105:18490–5.
Article7. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012; 149:274–93.
Article8. Hu JL, Hua YJ, Chen Y, Yu B, Gao S. Structural analysis of tumor-related single amino acid mutations in human MxA protein. Chin J Cancer. 2015; 34:583–93.
Article9. Haller O, Staeheli P, Schwemmle M, Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015; 23:154–63.
Article10. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010; 140:805–20.
Article11. Lee HJ, Kim JY, Song IH, Park IA, Yu JH, Ahn JH, et al. High mobility group B1 and N1 (HMGB1 and HMGN1) are associated with tumor-infiltrating lymphocytes in HER2-positive breast cancers. Virchows Arch. 2015; 467:701–9.
Article12. Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007; 6:975–90.
Article13. Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015; 15:231–42.
Article14. Simmons DP, Wearsch PA, Canaday DH, Meyerson HJ, Liu YC, Wang Y, et al. Type I IFN drives a distinctive dendritic cell maturation phenotype that allows continued class II MHC synthesis and antigen processing. J Immunol. 2012; 188:3116–26.
Article15. Minn AJ. Interferons and the immunogenic effects of cancer therapy. Trends Immunol. 2015; 36:725–37.
Article16. Calmon MF, Rodrigues RV, Kaneto CM, Moura RP, Silva SD, Mota LD, et al. Epigenetic silencing of CRABP2 and MX1 in head and neck tumors. Neoplasia. 2009; 11:1329–39.
Article17. Mushinski JF, Nguyen P, Stevens LM, Khanna C, Lee S, Chung EJ, et al. Inhibition of tumor cell motility by the interferon-inducible GTPase MxA. J Biol Chem. 2009; 284:15206–14.
Article18. Brown SG, Knowell AE, Hunt A, Patel D, Bhosle S, Chaudhary J. Interferon inducible antiviral MxA is inversely associated with prostate cancer and regulates cell cycle, invasion and docetaxel induced apoptosis. Prostate. 2015; 75:266–79.
Article19. Croner RS, Sturzl M, Rau TT, Metodieva G, Geppert CI, Naschberger E, et al. Quantitative proteome profiling of lymph node-positive vs. -negative colorectal carcinomas pinpoints MX1 as a marker for lymph node metastasis. Int J Cancer. 2014; 135:2878–86.20. Huber M, Bahr I, Kratzschmar JR, Becker A, Muller EC, Donner P, et al. Comparison of proteomic and genomic analyses of the human breast cancer cell line T47D and the antiestrogen-resistant derivative T47D-r. Mol Cell Proteomics. 2004; 3:43–55.
Article21. Becker M, Sommer A, Kratzschmar JR, Seidel H, Pohlenz HD, Fichtner I. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther. 2005; 4:151–68.22. Johansson HJ, Sanchez BC, Forshed J, Stal O, Fohlin H, Lewensohn R, et al. Proteomics profiling identify CAPS as a potential predictive marker of tamoxifen resistance in estrogen receptor positive breast cancer. Clin Proteomics. 2015; 12:8.
Article23. Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014; 20:1301–9.
Article24. Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009; 15:68–74.
Article25. Goc J, Germain C, Vo-Bourgais TK, Lupo A, Klein C, Knockaert S, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014; 74:705–15.
Article26. Martinet L, Garrido I, Filleron T, Le Guellec S, Bellard E, Fournie JJ, et al. Human solid tumors contain high endothelial venules: association with T- and B-lymphocyte infiltration and favorable prognosis in breast cancer. Cancer Res. 2011; 71:5678–87.
Article27. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.
Article28. Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999; 17:1474–81.
Article29. Lee HJ, Park IA, Park SY, Seo AN, Lim B, Chai Y, et al. Two histopathologically different diseases: hormone receptor-positive and hormone receptor-negative tumors in HER2-positive breast cancer. Breast Cancer Res Treat. 2014; 145:615–23.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of T-Lymphocyte Markers in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer
- Diagnosis and Treatment of HER2-Positive Breast Cancer
- Human Epidermal Growth Factor Receptor 2-positive Mucinous Carcinoma with Signet Ring Cell Differentiation, Which Showed Complete Response after Neoadjuvant Chemotherapy
- Prognostic Significance of Fibrotic Focus and Tumor Infiltrating Lymphocytes in Breast Cancer According to Molecular Subtypes
- Current Issues and Clinical Evidence in Tumor-Infiltrating Lymphocytes in Breast Cancer