J Korean Med Sci.
2016 Jun;31(6):836-842. 10.3346/jkms.2016.31.6.836.
Silencing of ABCG2 by MicroRNA-3163 Inhibits Multidrug Resistance in Retinoblastoma Cancer Stem Cells
- Affiliations
-
- 1Department of Ophthalmology, Linzi District People's Hospital, Zibo City, Shandong Province, P. R. China. jiaming_20150401@163.com
- KMID: 2373690
- DOI: http://doi.org/10.3346/jkms.2016.31.6.836
Abstract
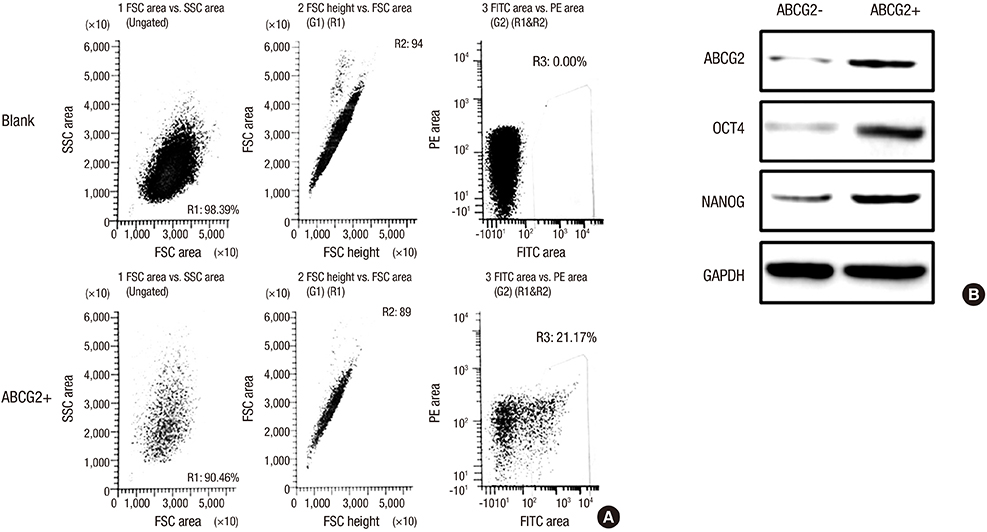
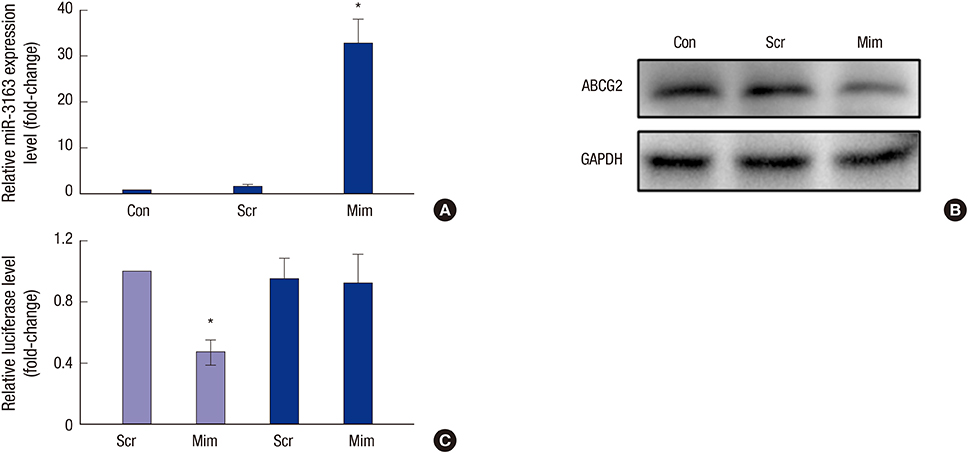
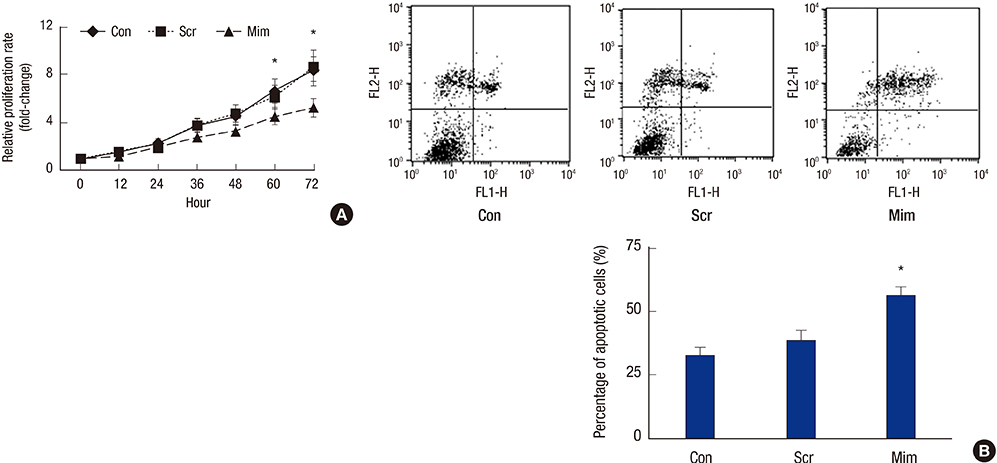
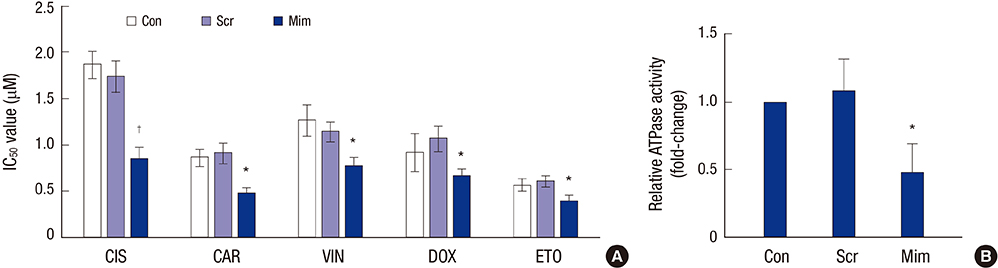
- To investigate the function and regulation mechanism of ATP-binding cassette, subfamily G, member 2 (ABCG2) in retinoblastoma cancer stem cells (RCSCs), a long-term culture of RCSCs from WERI-Rb1 cell line was successfully established based on the high expression level of ABCG2 on the surface of RCSCs. To further explore the molecular mechanism of ABCG2 on RCSCs, a microRNA that specifically targets ABCG2 was predicted. Subsequently, miR-3163 was selected and confirmed as the ABCG2-regulating microRNA. Overexpression of miR-3163 led to a significant decrease in ABCG2 expression. Additionally, ABCG2 loss-of-function induced anti-proliferation and apoptosis-promoting functions in RCSCs, and multidrug resistance to cisplatin, carboplatin, vincristine, doxorubicin, and etoposide was greatly improved in these cells. Our data suggest that miR-3163 has a significant impact on ABCG2 expression and can influence proliferation, apoptosis, and drug resistance in RCSCs. This work may provide new therapeutic targets for retinoblastoma.
Keyword
MeSH Terms
-
3' Untranslated Regions
ATP Binding Cassette Transporter, Sub-Family G, Member 2/antagonists & inhibitors/genetics/*metabolism
Antagomirs/metabolism
Antineoplastic Agents/toxicity
Apoptosis/drug effects
Base Sequence
Cell Line, Tumor
Cell Proliferation/drug effects
Drug Resistance, Neoplasm/drug effects
Gene Silencing
Humans
MicroRNAs/antagonists & inhibitors/genetics/*metabolism
Neoplasm Proteins/antagonists & inhibitors/genetics/*metabolism
Neoplastic Stem Cells/*metabolism
Retinoblastoma/metabolism/pathology
Sequence Alignment
Transfection
3' Untranslated Regions
ATP Binding Cassette Transporter, Sub-Family G, Member 2
Antagomirs
Antineoplastic Agents
MicroRNAs
Neoplasm Proteins
Figure
Reference
-
1. Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971; 68:820–823.2. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003; 100:3983–3988.3. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004; 432:396–401.4. Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005; 65:10946–10951.5. Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007; 445:111–115.6. Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007; 13:823–832.7. Seigel GM, Campbell LM, Narayan M, Gonzalez-Fernandez F. Cancer stem cell characteristics in retinoblastoma. Mol Vis. 2005; 11:729–737.8. Balla MM, Vemuganti GK, Kannabiran C, Honavar SG, Murthy R. Phenotypic characterization of retinoblastoma for the presence of putative cancer stem-like cell markers by flow cytometry. Invest Ophthalmol Vis Sci. 2009; 50:1506–1514.9. Zhong X, Li Y, Peng F, Huang B, Lin J, Zhang W, Zheng J, Jiang R, Song G, Ge J. Identification of tumorigenic retinal stem-like cells in human solid retinoblastomas. Int J Cancer. 2007; 121:2125–2131.10. Doyle L, Ross DD. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene. 2003; 22:7340–7358.11. Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998; 95:15665–15670.12. Diestra JE, Condom E, Del Muro XG, Scheffer GL, Pérez J, Zurita AJ, Muñoz-Seguí J, Vigués F, Scheper RJ, Capellá G, et al. Expression of multidrug resistance proteins P-glycoprotein, multidrug resistance protein 1, breast cancer resistance protein and lung resistance related protein in locally advanced bladder cancer treated with neoadjuvant chemotherapy: biological and clinical implications. J Urol. 2003; 170:1383–1387.13. Xie J, Jin B, Li DW, Shen B, Cong N, Zhang TZ. Dong P. ABCG2 regulated by MAPK pathways is associated with cancer progression in laryngeal squamous cell carcinoma. Am J Cancer Res. 2014; 4:698–709.14. Tsunoda S, Okumura T, Ito T, Kondo K, Ortiz C, Tanaka E, Watanabe G, Itami A, Sakai Y, Shimada Y. ABCG2 expression is an independent unfavorable prognostic factor in esophageal squamous cell carcinoma. Oncology. 2006; 71:251–258.15. Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008; 4:1–15.16. Miwa M, Tsukahara S, Ishikawa E, Asada S, Imai Y, Sugimoto Y. Single amino acid substitutions in the transmembrane domains of breast cancer resistance protein (BCRP) alter cross resistance patterns in transfectants. Int J Cancer. 2003; 107:757–763.17. Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, Robey R, Pommier Y, Fojo T, Bates SE. Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999; 59:5938–5946.18. Song J, Chang I, Chen Z, Kang M, Wang CY. Characterization of side populations in HNSCC: highly invasive, chemoresistant and abnormal Wnt signaling. PLoS One. 2010; 5:e11456.19. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233.20. Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355.21. Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010; 245:378–393.22. Gomez A, Ingelman-Sundberg M. Epigenetic and microRNA-dependent control of cytochrome P450 expression: a gap between DNA and protein. Pharmacogenomics. 2009; 10:1067–1076.23. Wang F, Xue X, Wei J, An Y, Yao J, Cai H, Wu J, Dai C, Qian Z, Xu Z, et al. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer. 2010; 103:567–574.24. Guo L, Liu Y, Bai Y, Sun Y, Xiao F, Guo Y. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer. 2010; 46:1692–1702.25. Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010; 285:4415–4422.26. Mohan A, Kandalam M, Ramkumar HL, Gopal L, Krishnakumar S. Stem cell markers: ABCG2 and MCM2 expression in retinoblastoma. Br J Ophthalmol. 2006; 90:889–893.27. Abbott BL, Colapietro AM, Barnes Y, Marini F, Andreeff M, Sorrentino BP. Low levels of ABCG2 expression in adult AML blast samples. Blood. 2002; 100:4594–4601.28. Ee PL, He X, Ross DD, Beck WT. Modulation of breast cancer resistance protein (BCRP/ABCG2) gene expression using RNA interference. Mol Cancer Ther. 2004; 3:1577–1583.29. Xie N, Mou L, Yuan J, Liu W, Deng T, Li Z, Jing Y, Hu Z. Modulating drug resistance by targeting BCRP/ABCG2 using retrovirus-mediated RNA interference. PLoS One. 2014; 9:e103463.30. Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011; 585:2087–2099.31. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838.32. Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012; 7:e34150.33. Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA, Hayward NK. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS One. 2010; 5:e9685.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Differential Expression of Stem Cell Markers and Vascular Endothelial Growth Factor in Human Retinoblastoma Tissue
- Effect of 5-FU and MTX on the Expression of Drug-resistance Related Cancer Stem Cell Markers in Non-small Cell Lung Cancer Cells
- Alectinib (CH5424802) antagonizes ABCB1- and ABCG2-mediated multidrug resistance in vitro, in vivo and ex vivo
- 2,3,7,8-Tetrachlorodibenzo-P-Dioxin Induced Cell-Specific Drug Transporters With Acquired Cisplatin Resistance in Cisplatin Sensitive Cancer Cells
- Cancer Stem Cells in Brain Tumors and Their Lineage Hierarchy