Ann Lab Med.
2017 Jan;37(1):18-27. 10.3343/alm.2017.37.1.18.
Development and Evaluation of a Duplex Real-Time PCR Assay With a Novel Internal Standard for Precise Quantification of Plasma DNA
- Affiliations
-
- 1Department of Laboratory Medicine, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China. sypan@njmu.edu.cn
- 2National Key Clinical Department of Laboratory Medicine, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
- KMID: 2373609
- DOI: http://doi.org/10.3343/alm.2017.37.1.18
Abstract
- BACKGROUND
Circulating levels of cell-free DNA increase in many pathologic conditions. However, notable discrepancies in the quantitative analysis of cell-free DNA from a large number of laboratories have become a considerable pitfall, hampering its clinical application.
METHODS
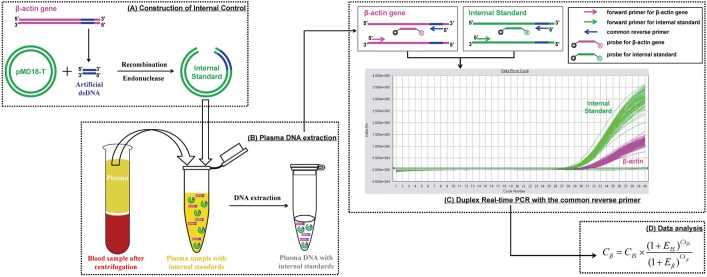
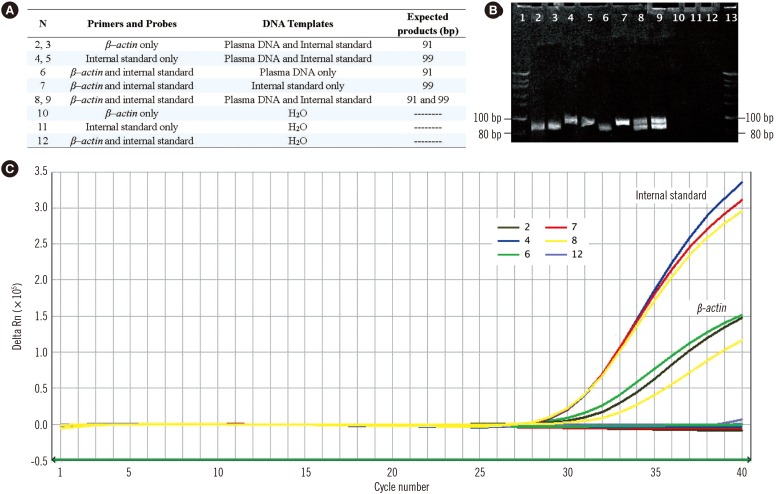
We designed a novel recombinant DNA fragment that could be applied as an internal standard in a newly developed and validated duplex real-time PCR assay for the quantitative analysis of total cell-free plasma DNA, which was tested in 5,442 healthy adults and 200 trauma patients.
RESULTS
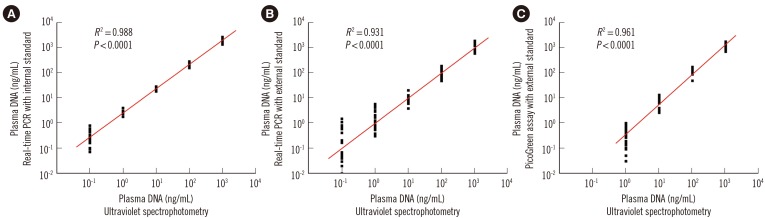
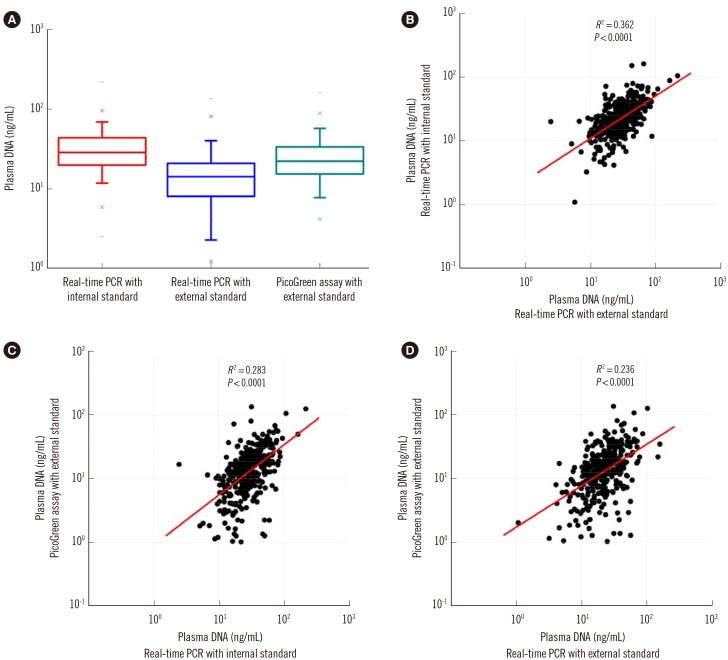
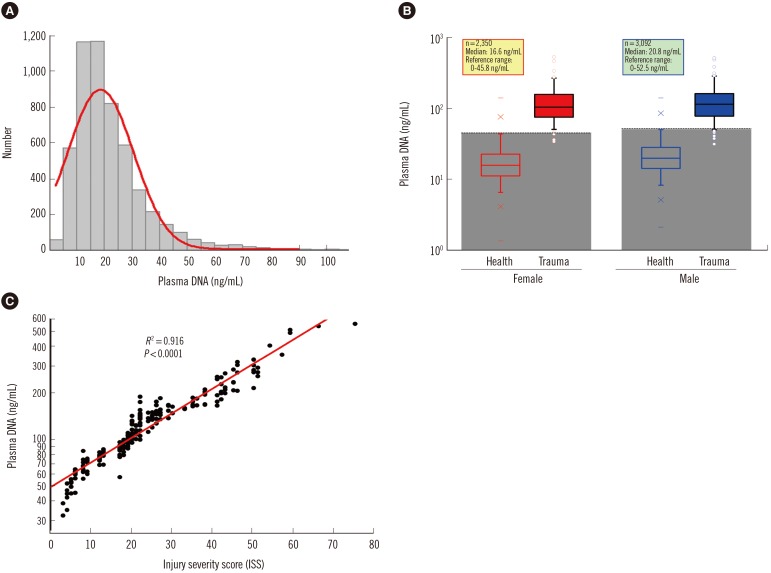
Compared with two traditional methods, this novel assay showed a lower detection limit of 0.1 ng/mL, lower intra- and inter-assay CVs, and higher accuracy in the recovery test. The median plasma DNA concentration of healthy males (20.3 ng/mL, n=3,092) was significantly higher than that of healthy females (16.1 ng/mL, n=2,350) (Mann-Whitney two-sample rank sum test, P<0.0001). The reference intervals of plasma DNA concentration were 0-45.8 ng/mL and 0-52.5 ng/mL for healthy females and males, respectively. The plasma DNA concentrations of the majority of trauma patients (96%) were higher than the upper normal cutoff values and were closely related to the corresponding injury severity scores (R²=0.916, P<0.0001).
CONCLUSIONS
This duplex real-time PCR assay with a new internal standard could eliminate variation and allow for more sensitive, repeatable, accurate, and stable quantitative measurements of plasma DNA, showing promising application in clinical diagnosis.
MeSH Terms
Figure
Reference
-
1. Mandel P, Metais P. Nucleic acids in plasma samples from human blood. C R Seances Soc Biol Fil. 1948; 142:241–243. PMID: 18875018.2. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977; 37:646–650. PMID: 837366.3. Tong YK, Lo YM. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta. 2006; 363:187–196. PMID: 16126188.4. Tissot C, Toffart AC, Villar S, Souquet PJ, Merle P, Moro-Sibilot D, et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J. 2015; 46:1773–1780. PMID: 26493785.5. Sozzi G, Roz L, Conte D, Mariani L, Andriani F, Lo Vullo S, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med. 2009; 179:69–74. PMID: 18787214.6. Altimari A, Grigioni AD, Benedettini E, Gabusi E, Schiavina R, Martinelli A, et al. Diagnostic role of circulating free plasma DNA detection in patients with localized prostate cancer. Am J Clin Pathol. 2008; 129:756–762. PMID: 18426736.7. Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, Kupis W, Rudzinski P, Langfort R, et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer. 2015; 113:476–483. PMID: 26125447.8. Paci M, Maramotti S, Bellesia E, Formisano D, Albertazzi L, Ricchetti T, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer. 2009; 64:92–97. PMID: 18804892.9. Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chassé E, et al. Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol. 2004; 22:4157–4164. PMID: 15483026.10. Skvortsova TE, Rykova EY, Tamkovich SN, Bryzgunova OE, Starikov AV, Kuznetsova NP, et al. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br J Cancer. 2006; 94:1492–1495. PMID: 16641902.11. Pinzani P, Salvianti F, Zaccara S, Massi D, De Giorgi V, Pazzagli M, et al. Circulating cell-free DNA in plasma of melanoma patients: qualitative and quantitative considerations. Clin Chim Acta. 2011; 412:2141–2145. PMID: 21839068.12. Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998; 62:768–775. PMID: 9529358.13. Manokhina I, Singh TK, Penaherrera MS, Robinson WP. Quantification of cell-free DNA in normal and complicated pregnancies: overcoming biological and technical issues. PLoS One. 2014; 9:e101500. PMID: 24987984.14. Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000; 46:319–323. PMID: 10702517.15. Rainer TH. Plasma DNA, prediction and post-traumatic complications. Clin Chim Acta. 2001; 313:81–85. PMID: 11694243.16. Saukkonen K, Lakkisto P, Varpula M, Varpula T, Voipio-Pulkki LM, Pettilä V, et al. Association of cell-free plasma DNA with hospital mortality and organ dysfunction in intensive care unit patients. Intensive Care Med. 2007; 33:1624–1627. PMID: 17541553.17. Chen JA, Meister S, Urbonaviciute V, Rodel F, Wilhelm S, Kalden JR, et al. Sensitive detection of plasma/serum DNA in patients with systemic lupus erythematosus. Autoimmunity. 2007; 40:307–310. PMID: 17516216.18. Macher HC, Suárez-Artacho G, Guerrero JM, Gomez-Bravo MA, Alvarez-Gómez S, Bernal-Bellido C, et al. Monitoring of transplanted liver health by quantification of organ-specific genomic marker in circulating DNA from receptor. PLoS One. 2014; 9:e113987. PMID: 25489845.19. Emons H, Corbisier P. Nucleic acid quantification-progress and pitfalls. Anal Bioanal Chem. 2014; 406:6469–6470. PMID: 25148828.20. Gormally E, Hainaut P, Caboux E, Airoldi L, Autrup H, Malaveille C, et al. Amount of DNA in plasma and cancer risk: a prospective study. Int J Cancer. 2004; 111:746–749. PMID: 15252845.21. de Kok JB, Hendriks JC, van Solinge WW, Willems HL, Mensink EJ, Swinkels DW. Use of real-time quantitative PCR to compare DNA isolation methods. Clin Chem. 1998; 44:2201–2204. PMID: 9761259.22. Jorgez CJ, Dang DD, Simpson JL, Lewis DE, Bischoff FZ. Quantity versus quality: optimal methods for cell-free DNA isolation from plasma of pregnant women. Genet Med. 2006; 8:615–619. PMID: 17079877.23. Szpechcinski A, Struniawska R, Zaleska J, Chabowski M, Orlowski T, Roszkowski K, et al. Evaluation of fluorescence-based methods for total vs. amplifiable DNA quantification in plasma of lung cancer patients. J Physiol Pharmacol. 2008; 59(S6):675–681. PMID: 19218694.24. Holdenrieder S, Stieber P, Chan LY, Geiger S, Kremer A, Nagel D, et al. Cell-free DNA in serum and plasma: comparison of ELISA and quantitative PCR. Clin Chem. 2005; 51:1544–1546. PMID: 16040855.25. Mauger F, Dulary C, Daviaud C, Deleuze JF, Tost J. Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem. 2015; 407:6873–6878. PMID: 26123439.26. Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014; 406:6499–6512. PMID: 24853859.27. Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003; 339:62–66. PMID: 12618301.28. Xue X, Teare MD, Holen I, Zhu YM, Woll PJ. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta. 2009; 404:100–104. PMID: 19281804.29. Zhang P, Ren J, Shen Z. A new quantitative method for circulating DNA level in human serum by capillary zone electrophoresis with laser-induced fluorescence detection. Electrophoresis. 2004; 25:1823–1828. PMID: 15213980.30. Fujii S, Inagaki K, Miyashita S, Nagasawa K, Chiba K, Takatsu A. A coupling system of capillary gel electrophoresis with inductively coupled plasma-mass spectrometry for the determination of double stranded DNA fragments. Metallomics. 2013; 5:424–428. PMID: 23604270.31. Camp CL, Sharp BL, Reid HJ, Entwisle J, Goenaga-Infante H. Analysis of mono-phosphate nucleotides as a potential method for quantification of DNA using high performance liquid chromatography-inductively coupled plasma-mass spectrometry. Anal Bioanal Chem. 2012; 402:367–372. PMID: 21877184.32. Huang CZ, Li KA, Tong SY. Determination of nucleic acids by a resonance light-scattering technique with alpha, beta, gamma, delta-tetrakis [4-(trimethylammoniumyl)phenyl]porphine. Anal Chem. 1996; 68:2259–2263. PMID: 9027231.33. Liu R, Yang J, Wu X, Sun C. Study on the resonance light scattering spectrum of berberine-cetyltrimethylammonium bromide system and the determination of nucleic acids at nanogram levels. Spectrochim Acta A Mol Biomol Spectrosc. 2002; 58:457–465. PMID: 11905531.34. Watts HJ, Yeung D, Parkes H. Real-time detection and quantification of DNA hybridization by an optical biosensor. Anal Chem. 1995; 67:4283–4289. PMID: 8633773.35. Pallisgaard N, Spindler KL, Andersen RF, Brandslund I, Jakobsen A. Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta. 2015; 446:141–146. PMID: 25896958.36. Wu TL, Zhang D, Chia JH, Tsao K, Sun CF, Wu JT. Cell-free DNA: measurement in various carcinoma and establishment of normal reference range. Clin Chim Acta. 2002; 321:77–87. PMID: 12031596.37. Lam NY, Rainer TH, Chan LY, Joynt GM, Lo YM. Time course of early and late changes in plasma DNA in trauma patients. Clin Chem. 2003; 49:1286–1291. PMID: 12881444.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Influence of Standard Curves on Relative Quantification using Real-time PCR
- Comparison between Real-Time PCR and Agarose Gel Electrophoresis for DNA Quantification
- Performance Evaluation of the Real-Q Cytomegalovirus (CMV) Quantification Kit Using Two Real-Time PCR Systems for Quantifying CMV DNA in Whole Blood
- Performance Evaluation of Real-Q HBV Quantification Kit for HBV DNA by Real-Time PCR
- Performance of the Real-Q EBV Quantification Kit for Epstein-Barr Virus DNA Quantification in Whole Blood