Tuberc Respir Dis.
2011 Jul;71(1):8-14.
MicroRNA-23a: A Novel Serum Based Diagnostic Biomarker for Lung Adenocarcinoma
- Affiliations
-
- 1Department of Internal Medicine, Konyang University Hospital, Konyang University College of Medicine, Daejeon, Korea. sk1609@hanmail.net
- 2Department of Laboratory Medicine, Konyang University Hospital, Konyang University College of Medicine, Daejeon, Korea.
- 3Myunggok Research Institute for Medical Science, Konyang University Hospital, Konyang University College of Medicine, Daejeon, Korea.
- 4Department of Internal Medicine, Kyungpook National University Hospital, Kyungpook National University College of Medicine, Daegu, Korea.
- 5Department of Pharmacology, Konyang University College of Medicine, Daejeon, Korea.
Abstract
- BACKGROUND
MicroRNAs (miRNAs) have demonstrated their potential as biomarkers for lung cancer diagnosis. In recent years, miRNAs have been found in body fluids such as serum, plasma, urine and saliva. Circulating miRNAs are highly stable and resistant to RNase activity along with, extreme pH and temperatures in serum and plasma. In this study, we investigated serum miRNA profiles that can be used as a diagnostic biomarker of non-small cell lung cancer (NSCLC).
METHODS
We compared the expression profile of miRNAs in the plasma of patients diagnosed with lung cancer using an miRNA microarray. The data from this assay were validated by quantitative real-time PCR (qRT-PCR).
RESULTS
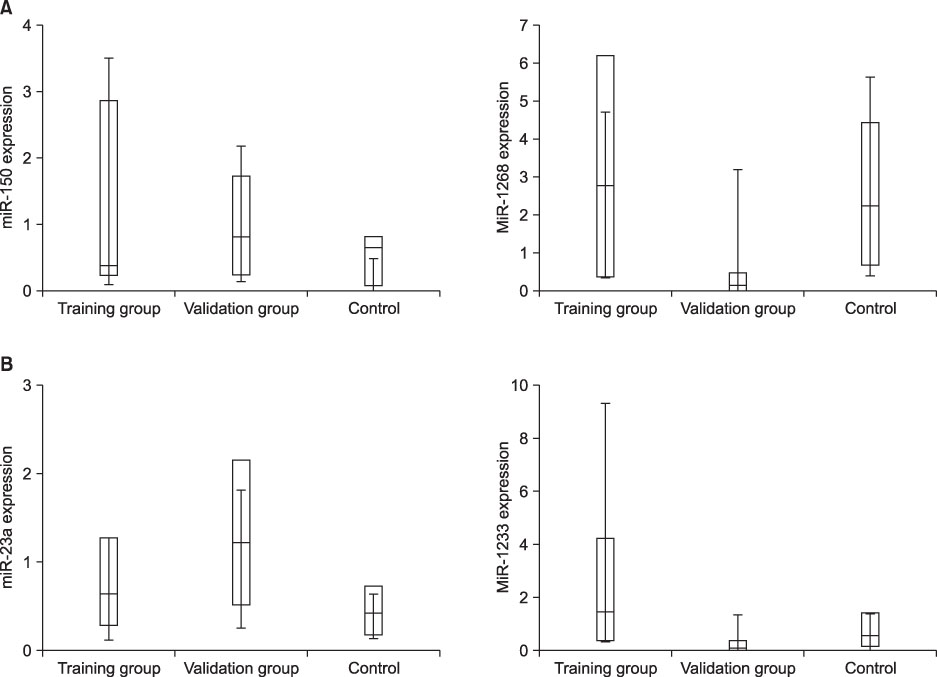
Six miRNAs were overexpressed and three miRNAs were underexpressed in both tissue and serum from squamous cell carcinoma (SCC) patients. Sixteen miRNAs were overexpressed and twenty two miRNAs were underexpressed in both tissue and serum from adenocarcinoma (AC) patients. Of the four miRNAs chosen for qRT-PCR analysis, the expression of miR-23a was consistent with microarray results from AC patients. Receiver operating characteristic (ROC) curve analyses were done and revealed that the level of serum miR-23a was a potential marker for discriminating AC patients from chronic obstructive pulmonary disease (COPD) patients.
CONCLUSION
Although a small number of patients were examined, the results from our study suggest that serum miR-23a can be used in the diagnosis of AC.
MeSH Terms
-
Adenocarcinoma
Biomarkers
Body Fluids
Carcinoma, Non-Small-Cell Lung
Carcinoma, Squamous Cell
Gene Expression Profiling
Humans
Hydrogen-Ion Concentration
Lung
Lung Neoplasms
MicroRNAs
Plasma
Pulmonary Disease, Chronic Obstructive
Real-Time Polymerase Chain Reaction
Ribonucleases
ROC Curve
Saliva
Adenocarcinoma
Lung Neoplasms
MicroRNAs
Ribonucleases
Figure
Reference
-
1. Ambros V. The functions of animal microRNAs. Nature. 2004. 431:350–355.2. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004. 116:281–297.3. Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006. 22:165–173.4. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006. 103:2257–2261.5. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006. 9:189–198.6. Son JW, Kim YJ, Cho HM, Lee SY, Jang JS, Choi JE, et al. MicroRNA expression profiles in Korean non-small cell lung cancer. Tuberc Respir Dis. 2009. 67:413–421.7. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005. 120:15–20.8. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008. 105:10513–10518.9. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010. 50:298–301.10. Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009. 4:e6229.11. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008. 141:672–675.12. Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006. 16:1289–1298.13. Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008. 26:462–469.14. Okies JE, Dietl C, Garrison HB, Starr A. Early and late results of resection of ventricular aneurysm. J Thorac Cardiovasc Surg. 1978. 75:255–260.15. Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009. 27:2030–2037.16. Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004. 64:3753–3756.17. Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011. 17:1875–1882.18. Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009. 45:2197–2206.19. Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008. 27:3845–3855.20. Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006. 5:24.21. Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2010 Nov 9 [Epub]. DOI: 10.1002/med.20215.22. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008. 18:997–1006.23. Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, et al. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009. 23:433–438.24. Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010. 38:7248–7259.25. Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010. 394:184–188.26. Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011. 50:136–142.27. Scapoli L, Palmieri A, Lo Muzio L, Pezzetti F, Rubini C, Girardi A, et al. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010. 23:1229–1234.28. Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med. 2011. 15:1593–1602.29. Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007. 25:387–392.30. Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, et al. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010. 277:3726–3734.31. Huang S, He X, Ding J, Liang L, Zhao Y, Zhang Z, et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008. 123:972–978.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MicroRNA-374a Expression as a Prognostic Biomarker in Lung Adenocarcinoma
- Serum miR-3620-3p as a Novel Biomarker for Ankylosing Spondylitis
- Diagnostic performance of microRNA-34a, let-7f and microRNA-31 in epithelial ovarian cancer prediction
- A Case of Hyperamylasemia Associated with Lung Adenocarcinoma
- Serum CEA in Non-Small Cell Lung Cancer Patients