Lab Med Online.
2016 Jul;6(3):165-170. 10.3343/lmo.2016.6.3.165.
Nucleic Acid Extraction for the Quantification of Cytomegalovirus and Epstein-Barr Virus
- Affiliations
-
- 1Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. sung@amc.seoul.kr
- 2Department of Laboratory Medicine, Center for Diagnostic Oncology and Hematologic Malignancy Branch, Division of Translational & Clinical Research II, National Cancer Center, Goyang, Korea.
- KMID: 2308732
- DOI: http://doi.org/10.3343/lmo.2016.6.3.165
Abstract
- BACKGROUND
Availability of an international standard will improve the standardization of quantitative PCR (qPCR) for cytomegalovirus (CMV) and Epstein-Barr virus (EBV); however, nucleic acid extraction methods may affect qPCR results. This study was designed to determine whether routine measurement of DNA concentration and purity is required in qPCR for CMV and EBV. In addition, the performance of the automated QIASymphony DSP DNA Mini kit (Qiagen, USA) and the manual QIAamp DNA Blood Mini kit (Qiagen) in extracting DNA from whole blood samples was compared.
METHODS
The concentration and purity of 300 extracted DNA samples were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, USA). A total of 72 and 54 whole blood samples were tested by artus CMV and EBV qPCR (Qiagen), respectively.
RESULTS
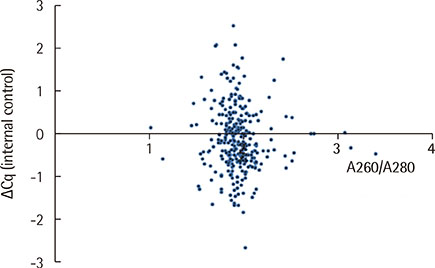
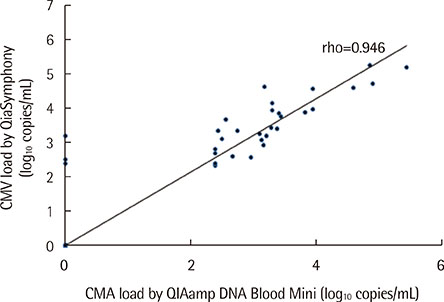
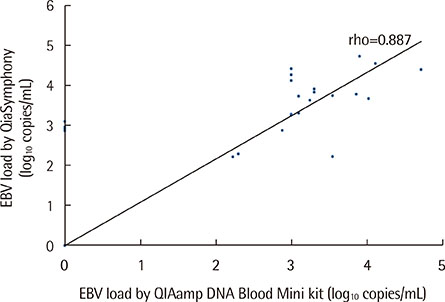
No correlation was found between DNA concentration and EBV DNA load or between DNA purity and the PCR inhibition measured by ΔCq (the difference between internal control Cq of the sample and that of the negative control). Quantification of CMV and EBV DNA using the two extraction methods showed highly similar results (rho=0.946 and 0.887, respectively). Of the 29 specimens that yielded CMV DNA by both methods, however, 8 specimens (27.6%) yielded higher CMV DNA loads with QIASymphony.
CONCLUSIONS
Routine measurement of DNA concentration and purity is not necessary for qPCR of CMV and EBV. The automated QIASymphony outperformed the manual QIAamp Blood Mini kit in extracting CMV and EBV DNA from whole blood samples.
Figure
Reference
-
1. Razonable RR, Emery VC. Management of CMV infection and disease in transplant patients. Herpes. 2004; 11:77–86.2. Cherikh WS, Kauffman HM, McBride MA, Maghirang J, Swinnen LJ, Hanto DW. Association of the type of induction immunosuppression with posttransplant lymphoproliferative disorder, graft survival, and patient survival after primary kidney transplantation. Transplantation. 2003; 76:1289–1293.
Article3. Baldanti F, Lilleri D, Gerna G. Monitoring human cytomegalovirus infection in transplant recipients. J Clin Virol. 2008; 41:237–241.
Article4. Gärtner B, Preiksaitis JK. EBV viral load detection in clinical virology. J Clin Virol. 2010; 48:82–90.
Article5. Hodinka RL. Human cytomegalovirus. In : Versalovic J, editor. Manual of clinical microbiology. 10th ed. Washington, DC: ASM Press;2011. p. 1558–1574.6. Gärtner BC. Epstein-Barr virus. In : Versalovic J, editor. Manual of clinical microbiology. 10th ed. Washington, DC: ASM Press;2011. p. 1575–1584.7. U.S. Food and Drug Administration. Nucleic acid based tests. Updated on Jan 2015. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm330711.htm.8. Fryer JF, Heath AB, Anderson R, Minor PD. the collaborative study group. Collaborative study to evaluate the proposed 1st WHO International Standard for human cytomegalovirus (HCMV) for nucleic acid amplification (NAT)-based assays. WHO ECBS Report 2010. WHO/BS/10.2138.9. Fryer JF, Heath AB, Wilkinson DE, Minor PD. the collaborative study group. Collaborative study to evaluate the proposed 1st WHO International Standard for Epstein-Barr virus (EBV) for nucleic acid amplification (NAT)-based assays. WHO ECBS Report 2011. WHO/BS/11.2172.10. Demeke T, Jenkins GR. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal Bioanal Chem. 2010; 396:1977–1990.
Article11. Clinical and Laboratory Standard Institute. Molecular diagnostic methods for infectious diseases. Approved guideline. MM03-A2. 2nd ed. Wayne, PA: Clinical and Laboratory Standard Institute;2006.12. Clinical and Laboratory Standard Institute. Quantitative molecular methods for infectious diseases. Approved guideline. MM6-A. Wayne, PA: Clinical and Laboratory Standard Institute;2003.13. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009; 55:611–622.
Article14. Kraft CS, Armstrong WS, Caliendo AM. Interpreting quantitative cytomegalovirus DNA testing: understanding the laboratory perspective. Clin Infect Dis. 2012; 54:1793–1797.
Article15. Kim S, Park SJ, Namgoong S, Sung H, Kim MN. Comparative evaluation of two automated systems for nucleic acid extraction of BK virus: NucliSens easyMAG versus BioRobot MDx. J Virol Methods. 2009; 162:208–212.
Article16. Lee KS, Park DS, Yoon KH, Lee YJ, Cho JH. Evaluation of ExiPrep16 automated system for the extraction of nucleic acids from nasopharyngeal swabs for the detection of respiratory viruses. Lab Med Online. 2013; 3:227–233.
Article17. Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, et al. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes. 2008; 1:70.
Article18. Riemann K, Adamzik M, Frauenrath S, Egensperger R, Schmid KW, Brockmeyer NH, et al. Comparison of manual and automated nucleic acid extraction from whole-blood samples. J Clin Lab Anal. 2007; 21:244–248.
Article19. Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006; 19:165–256.
Article20. Costa C, Mantovani S, Balloco C, Sidoti F, Fop F, Cavallo R. Comparison of two nucleic acid extraction and testing systems for HCMV-DNA detection and quantitation on whole blood specimens from transplant patients. J Virol Methods. 2013; 193:579–582.
Article21. Pillet S, Bourlet T, Pozzetto B. Comparative evaluation of the QIAsymphony RGQ system with the easyMAG/R-gene combination for the quantitation of cytomegalovirus DNA load in whole blood. Virol J. 2012; 9:231.
Article22. Bravo D, Clari MÀ, Costa E, Muñoz-Cobo B, Solano C, José Remigia M, et al. Comparative evaluation of three automated systems for DNA extraction in conjunction with three commercially available real-time PCR assays for quantitation of plasma Cytomegalovirus DNAemia in allogeneic stem cell transplant recipients. J Clin Microbiol. 2011; 49:2899–2904.
Article23. Gouarin S, Vabret A, Gault E, Petitjean J, Regeasse A, Hurault de Ligny B, et al. Quantitative analysis of HCMV DNA load in whole blood of renal transplant patients using real-time PCR assay. J Clin Virol. 2004; 29:194–201.
Article24. Vincent E, Gu Z, Morgenstern M, Gibson C, Pan J, Hayden RT. Detection of cytomegalovirus in whole blood using three different real-time PCR chemistries. J Mol Diagn. 2009; 11:54–59.
Article25. Forman M, Wilson A, Valsamakis A. Cytomegalovirus DNA quantification using an automated platform for nucleic acid extraction and real-time PCR assay setup. J Clin Microbiol. 2011; 49:2703–2705.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Automated Nucleic Acid Extraction Systems for Detecting Cytomegalovirus and Epstein-Barr Virus Using Real-Time PCR: A Comparison Study Between the QIAsymphony RGQ and QIAcube Systems
- Performance Evaluation of the ELITe InGenius System for Detecting Cytomegalovirus, EpsteinBarr Virus, and BK Virus Infections
- A Case of Epstein-Barr Virus Infection Presented as Evans Syndrome
- Performance of the Real-Q EBV Quantification Kit for Epstein-Barr Virus DNA Quantification in Whole Blood
- A Case of Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma Occurring in Thyroid Gland