J Bacteriol Virol.
2013 Sep;43(3):195-203. 10.4167/jbv.2013.43.3.195.
Variation and Characterization of Bacterial Communities Contaminating Two Saunas Operated at 64degrees C and 76degrees C
- Affiliations
-
- 1Department of Chemical and Biological Engineering, Seokyeong University, Seoul, Korea. baakdoo@skuniv.ac.kr
- KMID: 2286273
- DOI: http://doi.org/10.4167/jbv.2013.43.3.195
Abstract
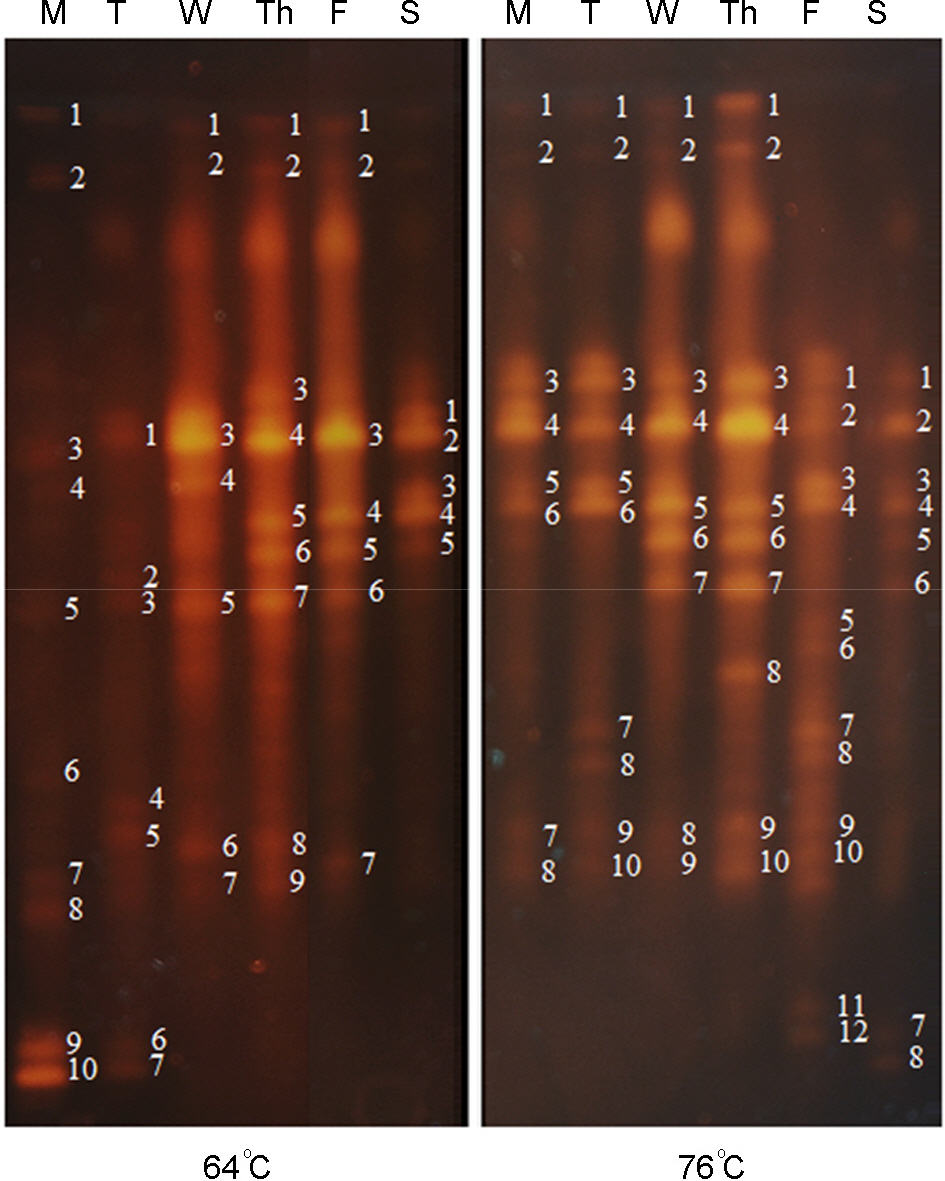
- This study was performed to analyze 6 day-term variations in bacterial communities contaminating the floor of two dry saunas that were operated at 64degrees C (low temp) and 76degrees C (high temp). Bacteria were sampled daily from the saunas for 6 days from Monday to Saturday. Genomic DNA was isolated directly from bacteria-collected cotton swabs. The diversity of the bacterial communities collected from the saunas was analyzed using thermal gradient gel electrophoresis (TGGE). The total numbers of DNA bands separated by TGGE for bacteria collected from the low temp and high temp sauna were 20 and 18, respectively, during the 6 days. Seven of 20 bacteria in the low temp sauna and eight of 18 bacteria in the high temp sauna were detected more than three times over the 6 experimental days. Twelve of the 26 bacterial genera contaminating the saunas were cross detected. Bacteria belonging to the genera Moraxella and Acinetobacter were selectively detected in the low temp sauna, whereas those belonging to Aquaspirillum, Chromobacterium, Aquabacterium, Gulbenkiania, Pelomonas, and Aquitalea were selectively detected in the high temp sauna. Three species of bacteria contaminating both the low and high temp saunas were thermophile or thermoduric. The results indicate that the sauna-contaminating bacteria may have been transferred from outside the saunas by user traffic but did not inhabit the saunas.
MeSH Terms
Figure
Reference
-
1). Lee JY, Park DH. Characterization of bacterial community contaminating floor of a hot and dry sauna. J Bacteriol Virol. 2012; 42:313–20.
Article2). Bott TL, Brock TD. Bacterial growth rates above 90°C in Yellowstone hot springs. Science. 1969; 164:1411–2.
Article3). Brock TD, Freeze H. Thermus aquaticus gen. n. and sp. n., a non-sporulating extreme thermophile. J Bacteriol. 1969; 98:289–97.4). Walsh C, Meade J, McGill K, Fanning S. The biodiversity of thermoduric bacteria isolated from whey. J Food Safe. 2012; 32:255–61.
Article5). Banykó J, Vyletelová M. Determining the source of Bacillus cereus and Bacillus licheniformis isolated from raw milk, pasteurised milk and yoghurt. Lett Appl Microbiol. 2009; 48:318–23.6). Brock TD, Boylen LK. Presence of thermophilic bacteria in laundry and domestic hot-water heaters. Appl Microbiol. 1973; 25:72–6.
Article7). Pask-Hughes R, Williama RA. Extremely thermophilic gram-negative bacteria from hot tap water. J Gen Microbil. 1975; 88:321–8.
Article8). Oshima T. Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. Nov. A nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol. 1974; 24:102–12.9). Ward J, Cockson A. Studies on a thermophilic bacillus: its isolation, properties, and temperature coefficient of growth. J Bacteriol. 1972; 112:1040–2.
Article10). Metzger WJ, Patterson R, Fink J, Semerdjian R, Roberts M. Sauna-takers disease. Hypersensitivity pneumonitis due to contaminated water in a home sauna. JAMA. 1976; 236:2209–11.
Article11). Scheldeman P, Pil A, Herman L, De Vos P, Heyndrickx M. Incidence and diversity of potentially highly heat-resistant spores isolated at dairy farms. Appl Environ Microbiol. 2005; 71:1480–94.
Article12). Murai R, Yoshida N. Geobacillus thermoglucosidasius endospores function as nuclei for the formation of single calcite crystals. Appl Environ Microbiol. 2013; 79:3085–90.13). Yokoya F, York GK. Effect of several environmental conditions on the “thermal death rate” of endospores of aerobic, thermophilic bacteria. Appl Microbiol. 1965; 13:993–9.
Article14). Martin PA, Travers RS. Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl Environ Microbiol. 1989; 55:2437–42.15). Stefanic P, Mandic-Mulec I. Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J Bacteriol. 2009; 191:1756–64.16). Cheung PY, Kinkle BK. Mycobacterium diversity and pyrene mineralization in petroleum-contaminated soils. Appl Environ Microbiol. 2001; 67:2222–9.17). Kimura H, Asada R, Masta A, Naganuma T. Distribution of microorganisms in the subsurface of the manus basin hydrothermal vent field in Papua New Guinea. Appl Environ Microbiol. 2003; 69:644–8.
Article18). Vary PS, Biedendieck R, Fuerch T, Meinhardt F, Rohde M, Deckwer WD, et al. Bacillus megaterium–from simple soil bacterium to industrial protein production host. Appl Microbiol Biotechnol. 2007; 76:957–67.19). Ferreira AC, Nobre MF, Rainey FA, Silva MT, Wait R, Burghardt J, et al. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int J Syst Bacteriol. 1997; 47:939–47.20). Scott RM. Bacterial endocarditis due to Neisseria flava. J Pediatr. 1971; 78:673–5.21). Schmidt B, Mulder IE, Musk CC, Aminov RI, Lewis M, Stokes CR, et al. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS One. 2011; 6:E28284.
Article22). Staley JT, Irgens RL, Brenner DJ. Enhydrobacter aerosaccus gen. nov., sp. nov., a gas-vacuolated, facultatively anaerobic, heterotrophic rod. Int J Syst Evol Microbiol. 1987; 37:289–91.23). Nishimura Y, Ino T, Lizuka H. Acinetobacter radioresistens sp. nov. isolated from cotteon and soil. Int J Syst Evol Microbiol. 1988; 38:209–11.24). Koburger JA, May SO. Isolation of Chromobacterium spp. from foods, soil, and water. Appl Environ Microbiol. 1982; 44:1463–5.25). Chen WM, Cho NT, Yang SH, Arun AB, Young CC, Sheu SY. Aquabacterium limnoticum sp. nov., isolated from a freshwater spring. Int J Syst Evol Microbiol. 2012; 62:698–704.26). Yoon JH, Kim IG, Oh TK. Acinetobacter marinus sp. nov. and Acinetobacter seohaensis sp. nov., isolated from sea water of the Yellow Sea in Korea. J Microbiol Biotechnol. 2007; 17:1743–50.27). Vaz-Moreira I, Nobre MF, Nunes OC, Manaia CM. Gulbenkiania mobilis gen. nov., sp. nov., isolated from treated municipal wastewater. Int J Syst Evol Microbiol. 2007; 57:1108–12.28). Gomila M, Bowien B, Falsen E, Moore ER, Lalucat J. Description of Pelomonas aquatica sp. nov. and Pelomonas puraquae sp. nov., isolated from industrial and haemodialysis water. Int J Syst Evol Microbiol. 2007; 57:2629–35.29). Nelson YM, Lion LW, Ghiorse WC, Shuler ML. Production of biogenic Mn oxides by Leptothrix discophora SS-1 in a chemically defined growth medium and evaluation of their Pb adsorption characteristics. Appl Environ Microbiol. 1999; 65:175–80.30). Lau HT, Faryna J, Triplett EW. Aquitalea magnusonii gen. Nov., sp. nov., a novel Gram-negative bacterium isolated from a humic lake. Int J Syst Evol Microbiol. 2006; 56:867–71.31). Gong XC, Liu ZS, Guo P, Chi CQ, Chen J, Wang XB, et al. Bacteria in crude oil survived autoclaving and stimulated differentially by exogenous bacteria. PLoS One. 2012; 7:e40842.
Article32). Loper JE, Lindow SE. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl Environ Microbiol. 1994; 60:1934–41.
Article33). Pearson HA. Rumen microbial ecology in mule deer. Appl Microbiol. 1969; 17:819–24.
Article34). Pernthaler J, Amann R. Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol Mol Biol Rev. 2005; 69:440–61.
Article35). Romero-Steiner S, Witek T, Balish E. Adherence of skin bacteria to human epithelial cells. J Clin Microbiol. 1990; 28:27–31.
Article36). Bowers RM, Sullivan AP, Costello EK, Collett JL Jr, Knight R, Fierer N. Source of bacteria in outdoor air across cities in the midwestern United State. Appl Environ Microbiol. 2011; 77:6350–6.37). Garcia SL, Jangid K, Whitman WB, Das KC. Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol. 2011; 102:7249–56.
Article38). Taylor WI, Schelhart D. Effect of temperature on transport and plating media for enteric pathogens. J Clin Microbiol. 1975; 2:281–6.
Article39). Ratkowsky DA, Olley J, McMeekin TA, Ball A. Relationship between temperature and growth of bacterial cultures. J Bacteriol. 1982; 149:1–5.40). Ratkowsky DA, Lowry RK, McMeekin TA, Stokes AN, Chandler RE. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J Bacteriol. 1983; 154:1222–6.
Article41). Nakasaki K, Sasaki M, Shoda M, Kubota H. Characteristics of mesophilic bacteria isolated during thermophilic composting of sewage sludge. Appl Environ Microbiol. 1985; 49:42–5.
Article42). Brock TD, Freeze H. Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J Bacteriol. 1969; 98:289–97.43). Verduin CM, Hol C, Fleer A, Van Dijk H, Van Belkum A. Moraxella catarrhalis: from emerging to established pathogen. Clin Microbiol Rev. 2002; 15:125–44.44). Vela AI, Arroyo E, Araqón V, Sánchez-Porro C, Latre MV, Cerdà-Cuéllar M, et al. Moraxella pluranimalium sp. nov., isolated from animal specimens. Int J Syst Evol Microbiol. 2009; 59:671–4.45). Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012; 22:850–9.
Article46). Morita RY. Psychrophilic bacteria. Bacteriol Rev. 1975; 39:144–67.
Article47). Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009; 459:950–6.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Characterization of Bacterial Community Contaminating Floor of A Hot and Dry Sauna
- Quantitative and Qualitative Estimation of Bacteria Contaminating Human Hairs
- Influence of Chemical- and Natural-Based Lotions on Bacterial Communities in Human Forearm Skin
- Intestinal Microbial Community Profiles of a Newborn Preterm Infant Using Pyrosequencing Analysis: Pilot Study
- Characterization of Soil Microorganism from Humus and Indigenous Microorganism Amendments