J Bacteriol Virol.
2013 Sep;43(3):186-194. 10.4167/jbv.2013.43.3.186.
A Virulent Salmonella enterica Serovar Enteritidis Phage SE2 with a Strong Bacteriolytic Activity of Planktonic and Biofilmed Cells
- Affiliations
-
- 1Department of Microbiology, Kyungpook National University, School of Medicine, Daegu, Korea. minkim@knu.ac.kr
- KMID: 2286272
- DOI: http://doi.org/10.4167/jbv.2013.43.3.186
Abstract
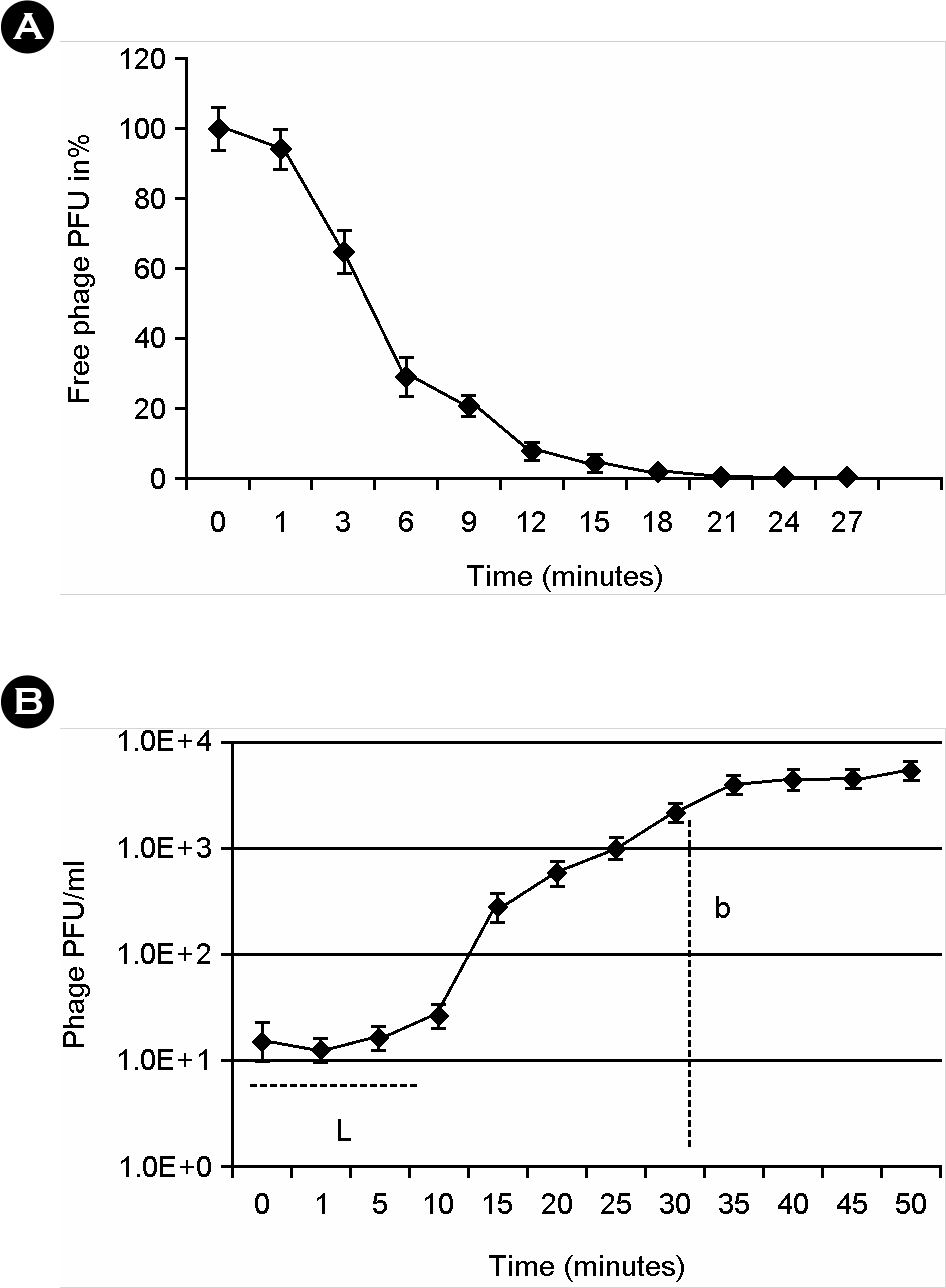
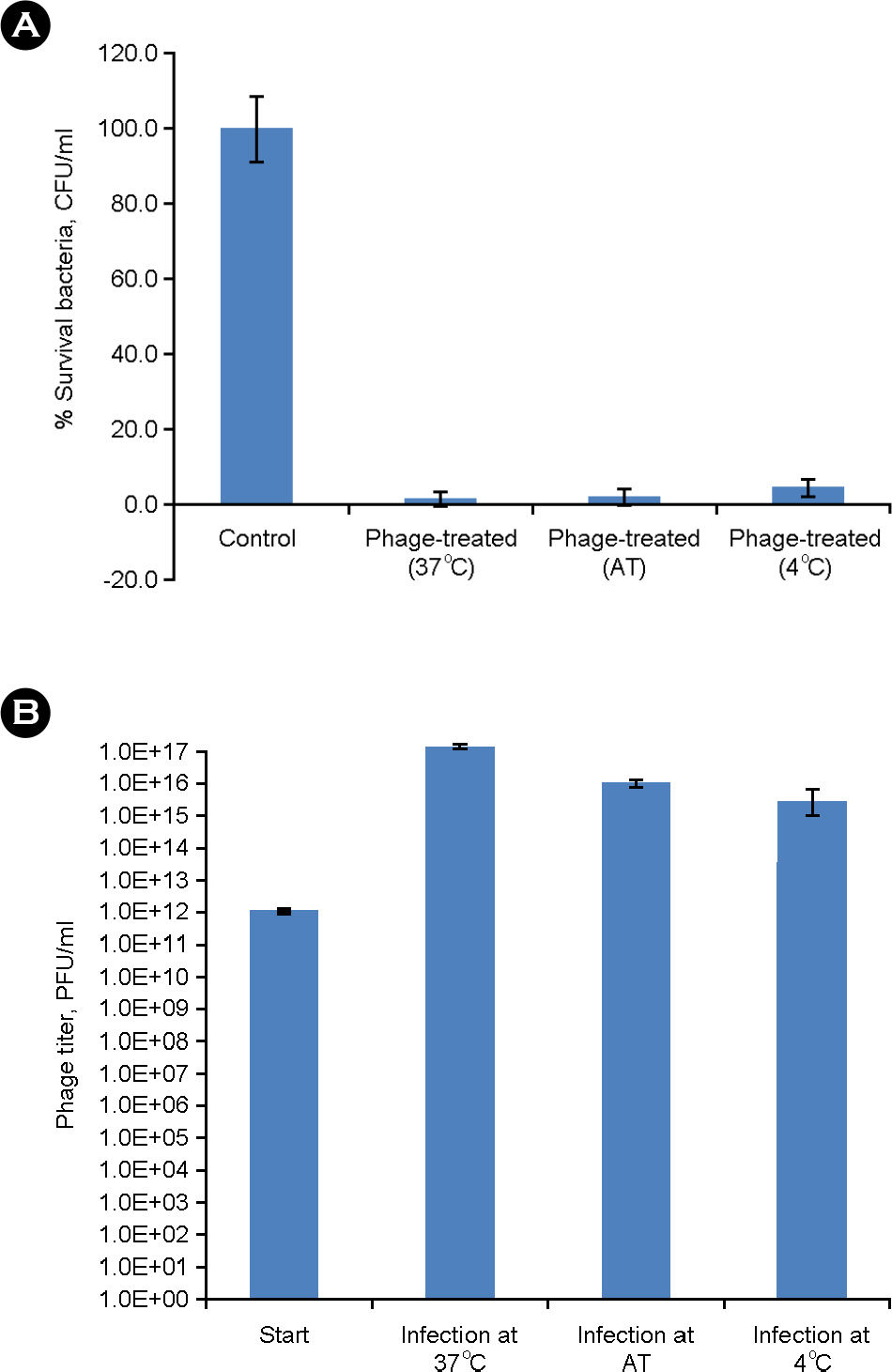
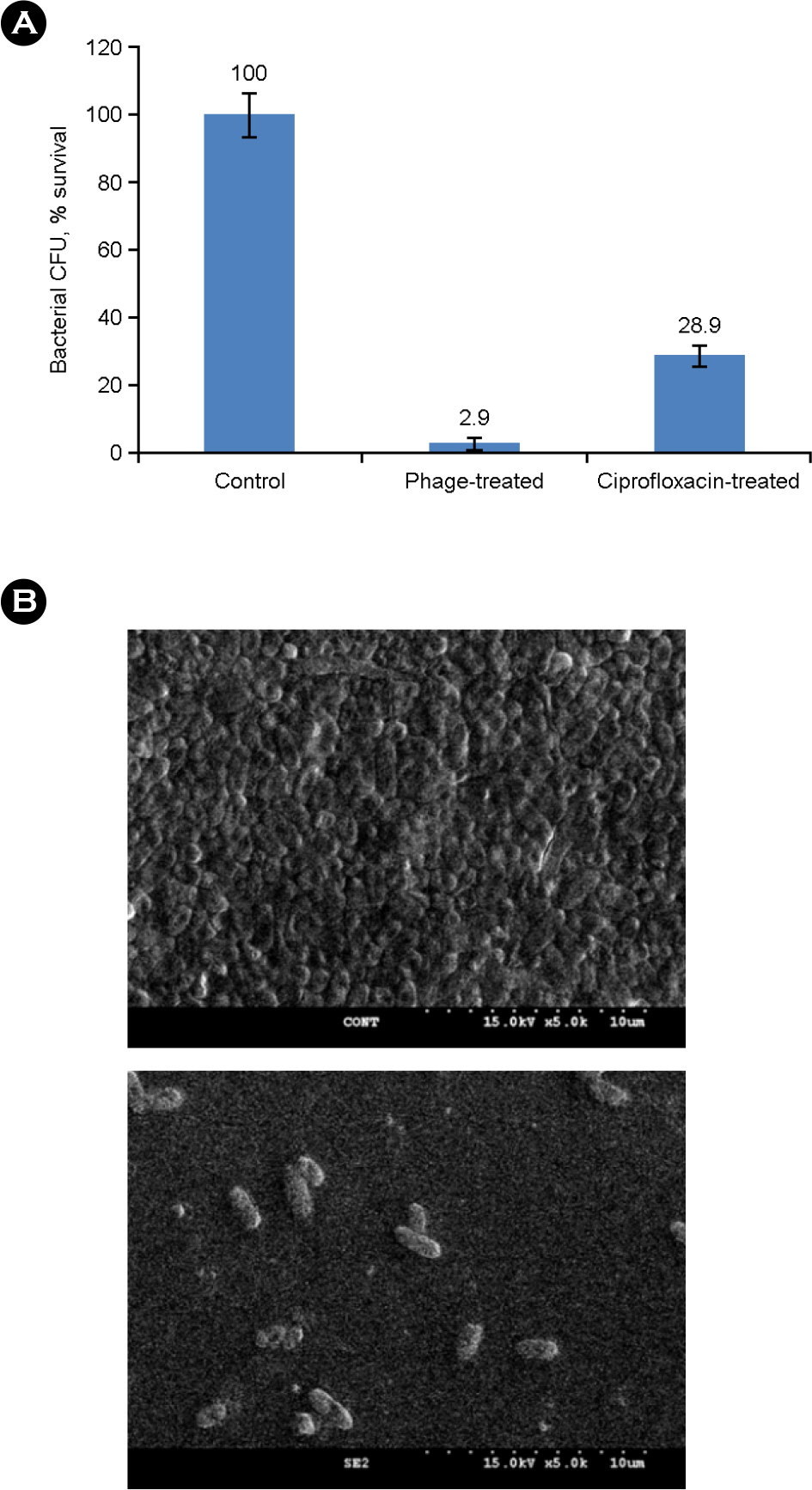
- Salmonella enterica serovar Enteritidis is one of the major food borne pathogens. Utilizing lytic bacteriophages against this pathogen can be a new and effective approach for the prevention of food-contamination and food-borne infection. In this study, we isolated and characterized a Salmonella Enteritidis specific lytic bacteriophage (phage SE2). The bacteriolytic activity of planktonic and biofilmed cells against an antibiotic resistant strain of Salmonella Enteritidis was also evaluated. Phage SE2 revealed an efficient bacteriolytic effect with biofilm dispersing ability and could maintain its virulence even at extreme pH and temperature. It can be a potential biotherapeutic agent against Salmonella Enteritidis.
MeSH Terms
Figure
Reference
-
1). Rodrigue DC, Tauxe RV, Rowe B. International increase in Salmonella enteritidis: A new pandemic? Epidemiol infect. 1990; 105:21–7.2). Guard-Petter J. The chicken, the egg and Salmonella Enteritidis. Environ Microbiol. 2001; 3:421–30.3). Aarestrup FM, Wiuff C, Mølbak K, Threlfall EJ. Is it time to change fluoroquinolone breakpoints for Salmonella spp.? Antimicrob Agents Chemother. 2003; 47:827–9.4). Kang ZW, Jung JH, Kim SH, Lee BK, Lee DY, Kim YJ, et al. Genotypic and Phenotypic Diversity of Salmonella Enteritidis Isolated from Chickens and Humans in Korea. J Vet Med Sci. 2009; 71:1433–8.5). Lindgren MM, Kotilainen P, Huovinen P, Hurme S, Lukinmaa S, Webber MA, et al. Reduced fluoroquinolone susceptibility in Salmonella enterica isolates from travelers, Finland. Emerg Infect Dis. 2009; 15:809–12.6). Tamang MD, Nam HM, Kim TS, Jang GC, Jung SC, Lim SK. Emergence of extended-spectrum β-Lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J Clin Microbiol. 2011; 49:2671–5.7). Kusumaningrum HD, Riboldi G, Hazeleger WC, Beumer RR. Survival of foodborne pathogens on stainless steel surfaces and cross-contamination to foods. Int J Food Microbiol. 2003; 85:227–36.
Article8). Chorianopoulos NG, Giaouris ED, Kourkoutas Y, Nychas GJ. Inhibition of the early stage of Salmonella enterica serovar Enteritidis biofilm development on stainless steel by cell-free supernatant of a Hafnia alvei culture. Appl Environ Microbiol. 2010; 76:2018–22.9). Solano C, García B, Valle J, Berasain C, Ghigo JM, Gamazo C, et al. Genetic analysis of Salmonella Enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002; 43:793–808.10). Soares VM, Pereira JG, Viana C, Izidoro TB, Bersot Ldos S, Pinto JP. Transfer of Salmonella Enteritidis to four types of surfaces after cleaning procedures and cross-contamination to tomatoes. Food Microbiol. 2012; 30:453–6.11). Van Houdt R, Michiels CW. Biofilm formation and the food industry, a focus on the bacterial outer surface. J Appl Microbiol. 2010; 109:1117–31.
Article12). Borysowski J, Weber-Dabrowska B, Górski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med. 2006; 231:366–77.
Article13). Goodridge LD, Bisha B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage. 2011; 1:130–7.
Article14). Mahony J, McAuliffe O, Ross RP, Van Sinderen D. Bacteriophages as biocontrol agents of food pathogens. Curr Opin Biotechnol. 2011; 22:157–63.
Article15). Andreatti Filho RL, Higgins JP, Higgins SE, Gaona G, Wolfenden AD, Tellez G, et al. Ability of bacteriophages isolated from different sources to reduce Salmonella enterica serovar Enteritidis in vitro and in vivo. Poult Sci. 2007; 86:1904–9.16). Atterbury RJ, Van Bergen MA, Ortiz F, Lovell MA, Harris JA, De Boer A, et al. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl Environ Microbiol. 2007; 73:4543–9.17). Goode D, Allen VM, Barrow PA. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl Environ Microbiol. 2003; 69:5032–6.18). Guenther S, Huwyler D, Richard S, Loessner MJ. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl Environ Microbiol. 2009; 75:93–100.19). Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother. 2010; 54:397–404.20). Rahman M, Kim S, Kim SM, Seol SY, Kim J. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling. 2011; 27:1087–93.21). Siringan P, Connerton PL, Payne RJ, Connerton IF. Bacteriophage-mediated dispersal of Campylobacter jejuni biofilms. Appl Environ Microbiol. 2011; 77:3320–6.22). Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007; 104:11197–202.
Article23). Tiwari BR, Kim S, Kim J. Complete genomic sequence of Salmonella enterica serovar Enteritidis phage SE2. J Virol. 2012; 86:7712.24). Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS one. 2009; 4:e4944.
Article25). Allison DG, Ruiz B, SanJose C, Jaspe A, Gilbert P. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol Lett. 1998; 167:179–84.26). Giaouris E, Chorianopoulos N, Nychas GJ. Effect of temperature, pH, and water activity of biofilm formation by Salmonella enterica Enteritidis PT4 on stainless steel surfaces as indicated by the bead vortexing method and conductance measurments. J Food Prot. 2005; 68:2149–54.27). Carey-Smith GV, Billington C, Cornelius AJ, Hudson JA, Heinemann JA. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol Lett. 2006; 258:182–6.28). O'Flynn G, Coffey A, Fitzgerald GF, Ross RP. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J Appl Microbiol. 2006; 101:251–9.29). Briandet R, Lacroix-Gueu P, Renault M, Lecart S, Meylheuc T, Bidnenko E, et al. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl Environ Microbiol. 2008; 74:2135–43.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Brain Death Caused by Salmonella Enterica Serovar Enteritidis Bacteremia
- Salmonella Serovars from Foodborne and Waterborne Diseases in Korea, 1998-2007: Total Isolates Decreasing Versus Rare Serovars Emerging
- Protection Against Salmonella Typhimurium, Salmonella Gallinarum, and Salmonella Enteritidis Infection in Layer Chickens Conferred by a Live Attenuated Salmonella Typhimurium Strain
- Characteristics of Epidemic Multidrugresistant Salmonella enterica Serovar Typhimurium DT104 Strains First Isolated in Korea
- A Comparison of Subtyping Methods for Differentiating
Salmonella enterica Serovar Enteritidis Isolates Obtained from Food and Human Sources