Allergy Asthma Immunol Res.
2015 May;7(3):265-275. 10.4168/aair.2015.7.3.265.
Copy Number Variation Burden on Asthma Subgenome in Normal Cohorts Identifies Susceptibility Markers
- Affiliations
-
- 1Genetics and Genomics Lab, Department of Studies in Zoology, University of Mysore, Manasagangotri, Karnataka, India. nallurbr@gmail.com
- 2Department of Pulmonology, JSS Hospital, Karnataka, India.
- KMID: 2260464
- DOI: http://doi.org/10.4168/aair.2015.7.3.265
Abstract
- PURPOSE
Asthma is a complex disease caused by interplay of genes and environment on the genome of an individual. Copy number variations (CNVs) are more common compared to the other variations that disrupt genome organization. The effect of CNVs on asthma subgenome has been less studied compared to studies on the other variations. We report the assessments of CNV burden in asthma genes of normal cohorts carried out in different geographical areas of the world and discuss the relevance of the observation with respect to asthma pathogenesis.
METHODS
CNV analysis was performed using Affymerix high-resolution arrays, and various bioinformatics tools were used to understand the influence of genes on asthma pathogenesis.
RESULTS
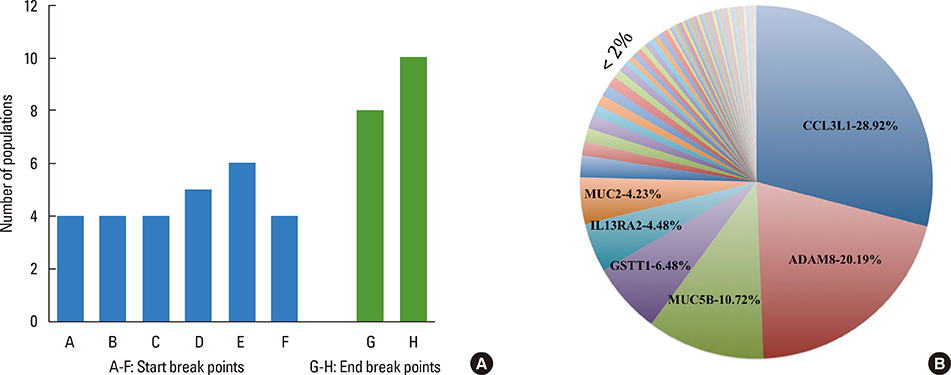
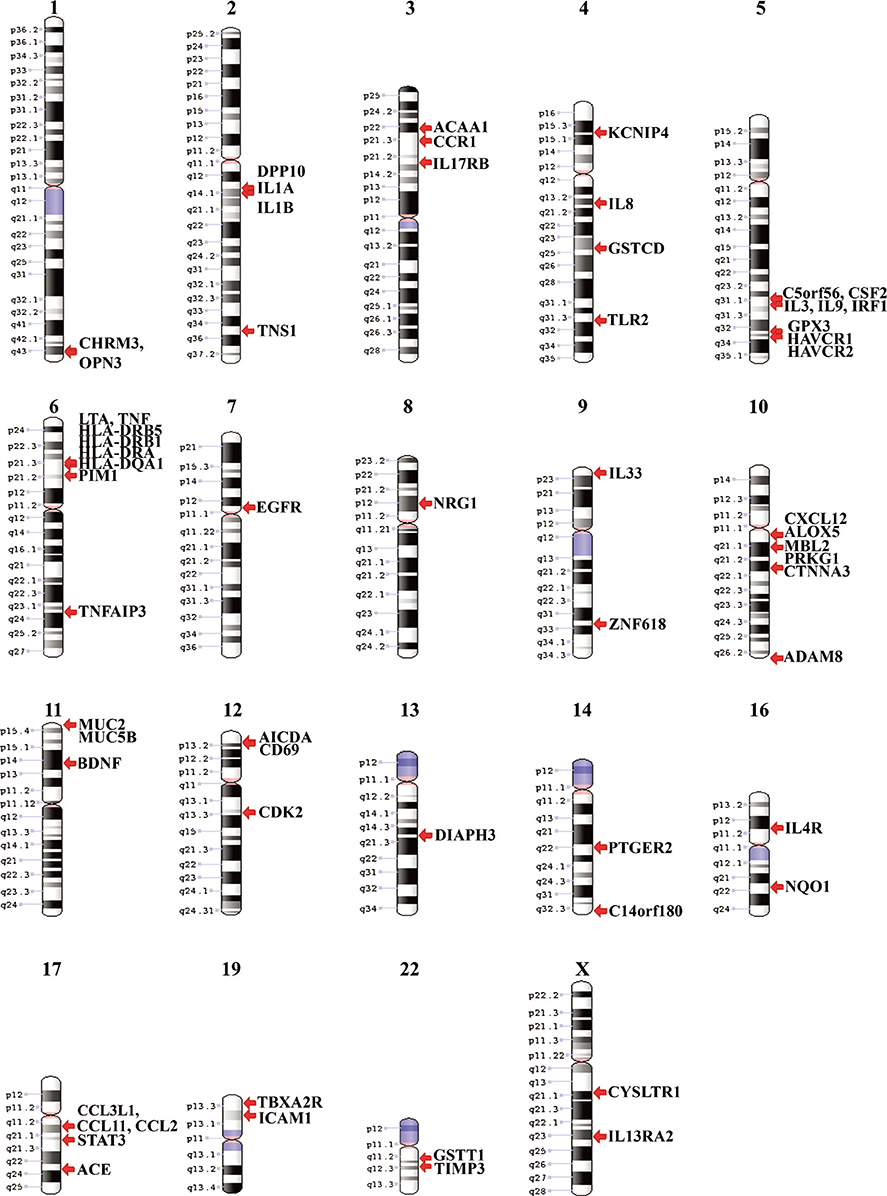
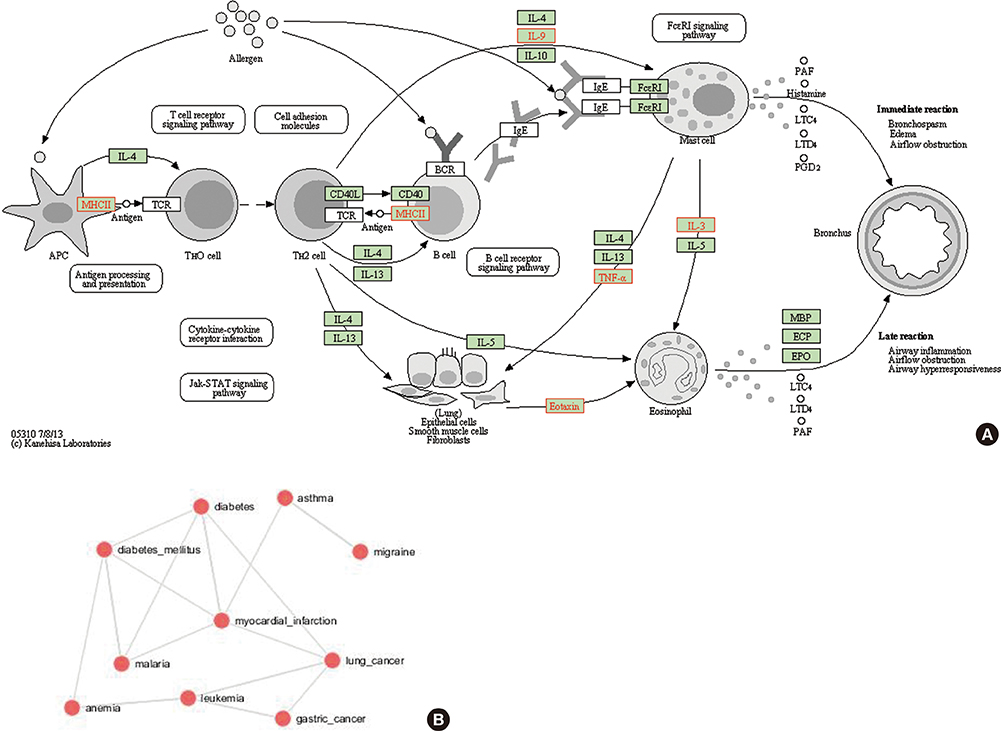
This study identified 61 genes associated with asthma and provided various mechanisms and pathways underlying asthma pathogenesis. CCL3L1, ADAM8, and MUC5B were the most prevalent asthma genes. Among them, CCL3L1 was found across all 12 populations in varying copy number states. This study also identified the inheritance of asthma-CNVs from parents to offspring creating the latent period for manifestation of asthma.
CONCLUSIONS
This study revealed CNV burden with varying copy number states and identified susceptibility towards the disease manifestation. It can be hypothesized that primary CNVs may not be the initiating event in the pathogenesis of asthma and additional preceding mutations or CNVs may be required. The initiator or primary CNVs sensitize normal cohorts leading to an increased probability of accumulating mutations or exposure to allergic stimulating agents that can augment the development of asthma.
MeSH Terms
Figure
Reference
-
1. Meng JF, Rosenwasser LJ. Unraveling the genetic basis of asthma and allergic diseases. Allergy Asthma Immunol Res. 2010; 2:215–227.2. International Union Against Tuberculosis and Lung Disease (FR). The global asthma report 2011. Paris: International Union Against Tuberculosis and Lung Disease;2011.3. Castro-Giner F, Kauffmann F, de Cid R, Kogevinas M. Gene-environment interactions in asthma. Occup Environ Med. 2006; 63:776–786. 7614. Hoffjan S, Ober C. Present status on the genetic studies of asthma. Curr Opin Immunol. 2002; 14:709–717.5. Park HS, Kim SH, Park CS. The role of novel genes in modifying airway responses in asthma. Curr Allergy Asthma Rep. 2006; 6:112–116.6. Zhang J, Paré PD, Sandford AJ. Recent advances in asthma genetics. Respir Res. 2008; 9:4.7. Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009; 93:22–26.8. Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007; 39:S48–S54.9. Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012; 63:35–61.10. Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007; 315:848–853.11. Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009; 10:451–481.12. Kitsiou-Tzeli S, Frysira H, Giannikou K, Syrmou A, Kosma K, Kakourou G, et al. Microdeletion and microduplication 17q21.31 plus an additional CNV, in patients with intellectual disability, identified by array-CGH. Gene. 2012; 492:319–324.13. Veerappa AM, Murthy MN, Vishweswaraiah S, Lingaiah K, Suresh RV, Nachappa SA, et al. Copy number variations burden on miRNA genes reveals layers of complexities involved in the regulation of pathways and phenotypic expression. PLoS One. 2014; 9:e90391.14. Rogers AJ, Chu JH, Darvishi K, Ionita-Laza I, Lehmann H, Mills R, et al. Copy number variation prevalence in known asthma genes and their impact on asthma susceptibility. Clin Exp Allergy. 2013; 43:455–462.15. International HapMap Consortium. The International HapMap Project. Nature. 2003; 426:789–796.16. Veerappa AM, Vishweswaraiah S, Lingaiah K, Murthy M, Manjegowda DS, Nayaka R, et al. Unravelling the complexity of human olfactory receptor repertoire by copy number analysis across population using high resolution arrays. PLoS One. 2013; 8:e66843.17. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14:128.18. Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41:W77–W83.19. Eichler EE. Copy number variation and human disease. Nat Educ. 2008; 1:1.20. Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011; 19:458–464.21. Mileyko Y, Joh RI, Weitz JS. Small-scale copy number variation and large-scale changes in gene expression. Proc Natl Acad Sci U S A. 2008; 105:16659–16664.22. Winchester L, Newbury DF, Monaco AP, Ragoussis J. Detection, breakpoint identification and detailed characterisation of a CNV at the FRA16D site using SNP assays. Cytogenet Genome Res. 2008; 123:322–332.23. Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008; 40:1253–1260.24. Cavalli-Sforza LL. Genes, peoples, and languages. Proc Natl Acad Sci U S A. 1997; 94:7719–7724.25. Lee H, Bae S, Choi BW, Choi JC, Yoon Y. Copy number variation of CCL3L1 influences asthma risk by modulating IL-10 expression. Clin Chim Acta. 2011; 412:2100–2104.26. Masoli M, Fabian D, Holt S, Beasley R. Global Initiative for Asthma (GINA) Program. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004; 59:469–478.27. Kosta K, Sabroe I, Goke J, Nibbs RJ, Tsanakas J, Whyte MK, et al. A Bayesian approach to copy-number-polymorphism analysis in nuclear pedigrees. Am J Hum Genet. 2007; 81:808–812.28. King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, et al. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol. 2004; 31:257–265.29. Holloway JW, Holgate ST. Identification and function of a Novel Candidate Gene for Asthma: ADAM 33. Allergol Int. 2005; 54:25–30.30. Mahesh PA. Unravelling the role of ADAM 33 in asthma. Indian J Med Res. 2013; 137:447–450.31. Paulissen G, Rocks N, Guéders MM, Bedoret D, Crahay C, Quesada-Calvo F, et al. ADAM-8, a metalloproteinase, drives acute allergen-induced airway inflammation. Eur J Immunol. 2011; 41:380–391.32. Domínguez-Luis M, Lamana A, Vazquez J, García-Navas R, Mollinedo F, Sánchez-Madrid F, et al. The metalloprotease ADAM8 is associated with and regulates the function of the adhesion receptor PSGL-1 through ERM proteins. Eur J Immunol. 2011; 41:3436–3442.33. Knolle MD, Owen CA. ADAM8: a new therapeutic target for asthma. Expert Opin Ther Targets. 2009; 13:523–540.34. Evans CM, Kim K, Tuvim MJ, Dickey BF. Mucus hypersecretion in asthma: causes and effects. Curr Opin Pulm Med. 2009; 15:4–11.35. Groneberg DA, Eynott PR, Lim S, Oates T, Wu R, Carlstedt I, et al. Expression of respiratory mucins in fatal status asthmaticus and mild asthma. Histopathology. 2002; 40:367–373.36. Morcillo EJ, Cortijo J. Mucus and MUC in asthma. Curr Opin Pulm Med. 2006; 12:1–6.37. Vinall LE, Fowler JC, Jones AL, Kirkbride HJ, de Bolós C, Laine A, et al. Polymorphism of human mucin genes in chest disease: possible significance of MUC2. Am J Respir Cell Mol Biol. 2000; 23:678–686.38. Nambiar M, Raghavan SC. Chromosomal translocations among the healthy human population: implications in oncogenesis. Cell Mol Life Sci. 2013; 70:1381–1392.39. Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007; 316:445–449.40. Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008; 82:477–488.41. Ehli EA, Abdellaoui A, Hu Y, Hottenga JJ, Kattenberg M, van Beijsterveldt T, et al. De novo and inherited CNVs in MZ twin pairs selected for discordance and concordance on Attention Problems. Eur J Hum Genet. 2012; 20:1037–1043.42. Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu Rev Immunol. 2004; 22:789–815.43. Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2010; 125:S73–S80.44. Robinson DS. The role of the T cell in asthma. J Allergy Clin Immunol. 2010; 126:1081–1091.45. Conroy DM, Williams TJ. Eotaxin and the attraction of eosinophils to the asthmatic lung. Respir Res. 2001; 2:150–156.46. Filipović M, Cekić S. The role of eosinophils in asthma. Facta Univ Ser Med Biol. 2001; 8:6–10.47. Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007; 104:8685–8690.48. Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, et al. De novo rates and selection of large copy number variation. Genome Res. 2010; 20:1469–1481.49. Walsh KM. Copy-number variation and the etiology of chronic diseases: computational detections, laboratory validations, and public health relevance [Dissertation]. New Haven (CT): Yale University;2011.50. Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012; 367:1321–1331.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical implications of copy number variations in autoimmune disorders
- Copy Number Variations in the Human Genome: Potential Source for Individual Diversity and Disease Association Studies
- Correlation of DEFA1 Gene Copy Number Variation with Intestinal Involvement in Behcet's Disease
- Association between Mitochondrial D-loop Polymorphism and Copy Number
- Web-Based Database and Viewer of East Asian Copy Number Variations