Korean J Hematol.
2006 Mar;41(1):1-7. 10.5045/kjh.2006.41.1.1.
Overexpression of the Fanconi Anemia A Gene in Hela and MCF10A Cells
- Affiliations
-
- 1Department of Physiology, Chonbuk National University Medical School, Jeonju, Korea. parkwh71@yahoo.co.kr
- KMID: 2252321
- DOI: http://doi.org/10.5045/kjh.2006.41.1.1
Abstract
-
BACKGROUND: Fanconi Anemia (FA) is an autosomal recessive inherited disease, which is characterized by developmental abnormalities, progressive bone marrow failure and a predisposition to cancer. The phenotypes of FA cells show extreme sensitivities towards oxygen and DNA cross linking agents, such as diepoxybutane and mitomycin C (MMC).
METHODS
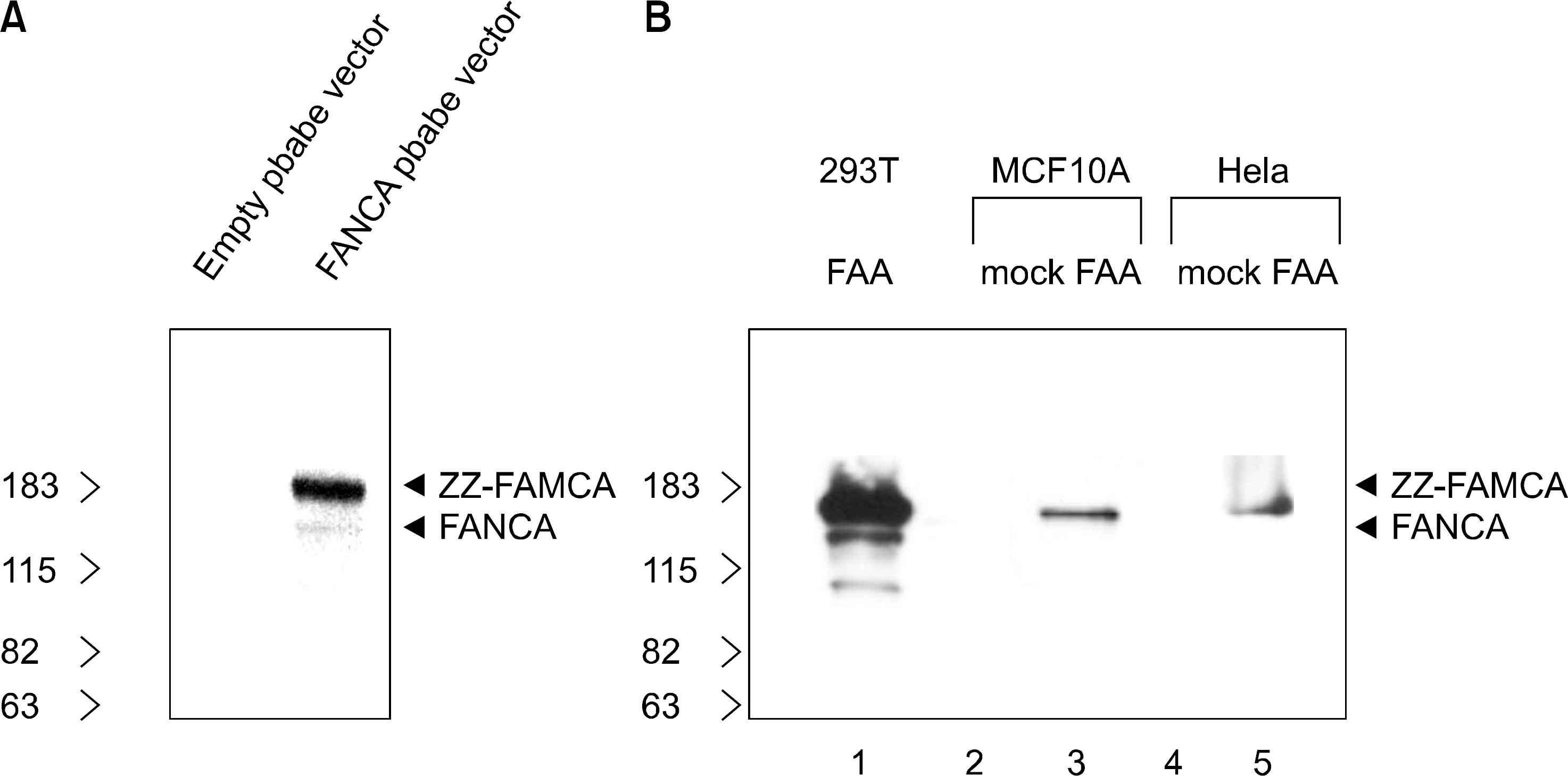
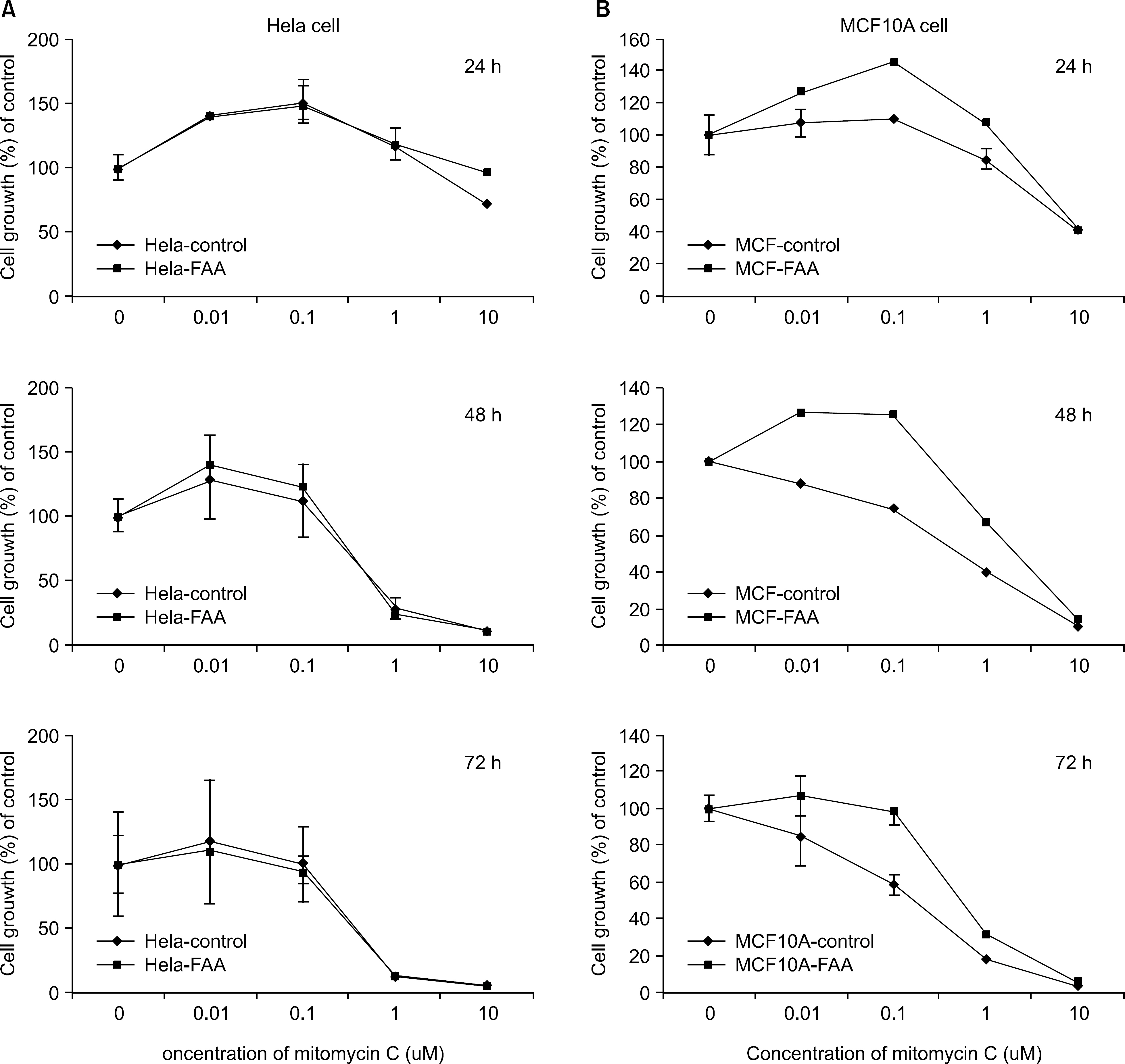
In the current study, retroviruses expressing the FANCA gene were prepared to create the stable cell lines, Hela (cervical carcinoma) and MCF10A (breast). The expression of FANCA protein in the Hela and MCF10A stable cells, following puromycin selection, was checked using Western blot. The difference in the cell growth between the parent and FANCA expressing cells following MMC treatment was checked using the MTT assay.
RESULTS
The expression of exogenous FANCA protein in the Hela and MCF10A stable cells was observed using Western blot. The MCF10A cells expressing exogenous FANCA were resistant to MMC concentrations with the range 0.01~1 micrometer compared with the MCF10 parent cells. However, at an MMC concentration of 10 micrometer, there was no difference in the susceptibility between the parent and FANCA expressing MCF10 cells. The Hela cells expressing FANCA showed no resistance at any MMC concentration (0.01~10 micrometer).
CONCLUSION
FANCA protein is an important factor for resistance to the cross linking agent, MMC, in MCF10A breast cells, but not in Hela cervical carcinoma cells.
Keyword
MeSH Terms
Figure
Reference
-
1). Bagby GC Jr. Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003. 10:68–76.
Article2). Joenje H., Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001. 2:446–57.
Article3). D'Andrea AD., Grompe M. The Fanconi anaemia/BR-CA pathway. Nat Rev Cancer. 2003. 3:23–34.4). Kwee ML., Poll EH., van de Kamp JJ., de Koning H., Eriksson AW., Joenje H. Unusual response to bifunctional alkylating agents in a case of Fanconi anaemia. Hum Genet. 1983. 64:384–7.
Article5). Joenje H., Oostra AB., Wijker M, et al. Evidence for at least eight Fanconi anemia genes. Am J Hum Genet. 1997. 61:940–4.
Article6). Meetei AR., de Winter JP., Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003. 35:165–70.
Article7). Meetei AR., Levitus M., Xue Y, et al. X-linked inhe ritance of Fanconi anemia complementation group B. Nat Genet. 2004. 36:1219–24.8). Medhurst AL., Huber PA., Waisfisz Q., de Winter JP., Mathew CG. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum Mol Genet. 2001. 10:423–4.
Article9). Garcia-Higuera I., Taniguchi T., Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001. 7:249–62.
Article10). Moynahan ME., Pierce AJ., Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001. 7:263–72.
Article11). Moynahan ME., Chiu JW., Koller BH., Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999. 4:511–8.
Article12). Lundberg R., Mavinakere M., Campbell C. Deficient DNA end joining activity in extracts from fanconi anemia fibroblasts. J Biol Chem. 2001. 276:9543–9.
Article13). Donahue SL., Campbell C. A DNA double strand break repair defect in Fanconi anemia fibroblasts. J Biol Chem. 2002. 277:46243–7.
Article14). Donahue SL., Campbell C. A Rad50-dependent pathway of DNA repair is deficient in Fanconi anemia fibroblasts. Nucleic Acids Res. 2004. 32:3248–57.
Article15). Pfeiffer P., Goedecke W., Kuhfittig-Kulle S., Obe G. Pathways of DNA double-strand break repair and their impact on the prevention and formation of chromosomal aberrations. Cytogenet Genome Res. 2004. 104:7–13.
Article16). van den Bosch M., Lohman PH., Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem. 2002. 383:873–92.
Article17). Folias A., Matkovic M., Bruun D, et al. BRCA1 interacts directly with the Fanconi anemia protein FAN-CA. Hum Mol Genet. 2002. 11:2591–7.
Article18). Howlett NG., Taniguchi T., Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002. 297:606–9.19). Hirsch B., Shimamura A., Moreau L, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004. 103:2554–9.
Article20). West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003. 4:435–45.
Article