Korean Circ J.
2011 Jul;41(7):385-393. 10.4070/kcj.2011.41.7.385.
Deoxyribonucleic Acid Copy Number Aberrations in Vasospastic Angina Patients Using an Array Comparative Genomic Hybridization
- Affiliations
-
- 1Cardiovascular Center and Cardiology Division, College of Medicine, The Catholic University of Korea, Seoul, Korea. hojheart@catholic.ac.kr
- 2Catholic Neuroscience Center, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 3Department of Pharmacology, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 2225102
- DOI: http://doi.org/10.4070/kcj.2011.41.7.385
Abstract
- BACKGROUND AND OBJECTIVES
Vasospastic angina (VA) is a specific type of coronary artery disease and develops as a result of coronary artery spasm. Recently, a few studies have revealed that VA caused by coronary artery spasm is related to genetic traits. The objective of this study was to use the recently developed technique of array comparative genomic hybridization (CGH) to screen the genetic aberrations of VA.
SUBJECTS AND METHODS
To identify candidate genes that might be causally involved in the pathogenesis of VA, genomic deoxyribonucleic acids were extracted from whole blood of 28 patients with VA who presented at Department of Cardiology at Seoul St. Mary's Hospital, Seoul, Korea. The copy number profiles of these patients was then analyzed using array CGH and reverse transcriptase (RT) quantitative polymerase chain reaction (PCR).
RESULTS
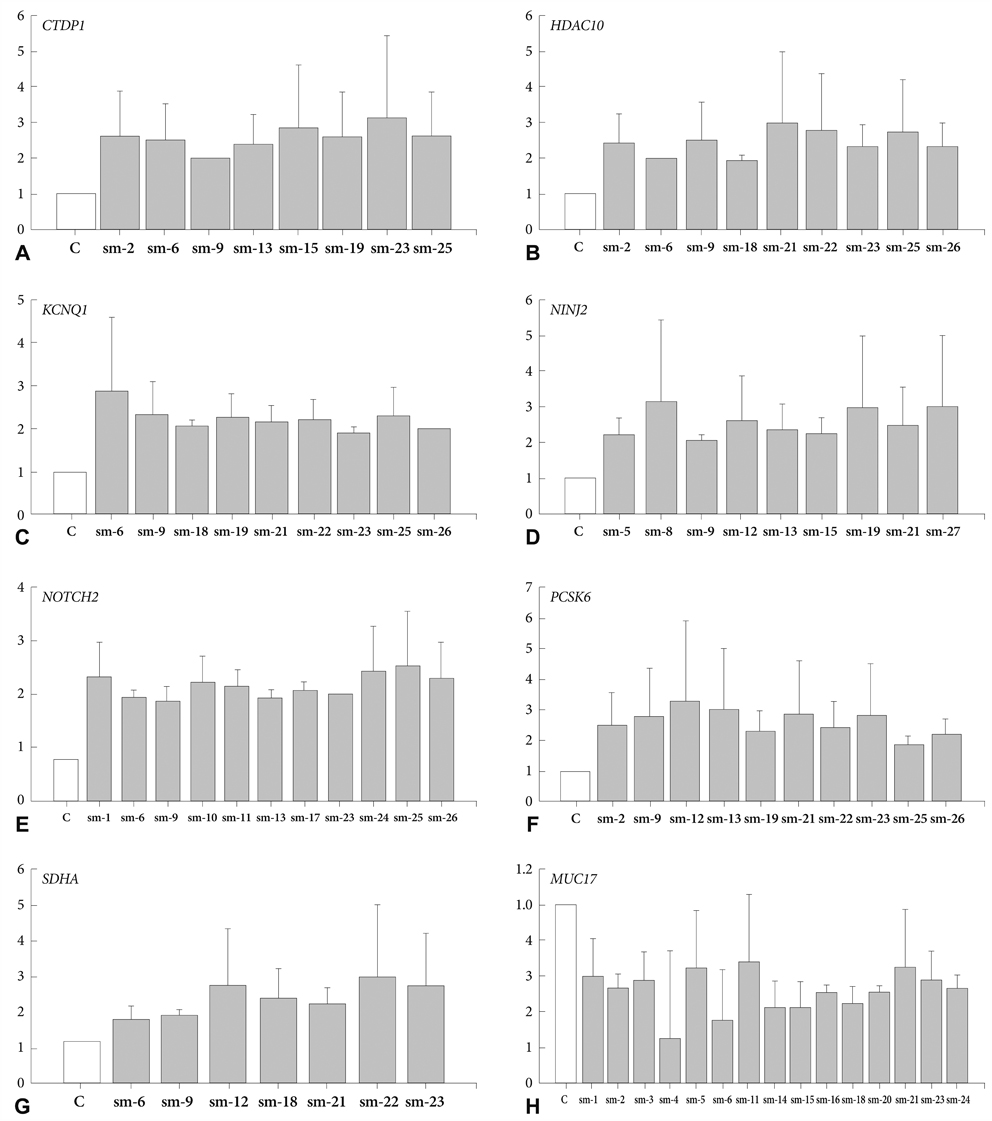
Array CGH revealed gains in 31 different regions, with losses in the 4q35.2, 7q22.1, 10q26.3, 15q11.2, 16p13.11, 17p11.2 and 19q13.3 regions (more than 32% aberration in these regions). Several loci were found to be frequently including gains of 5p and 11q (50% of samples). The most common losses were found in 7q (54% of samples). Copy number aberrations in chromosomal regions were detected and corresponding genes were confirmed by RT quantitative PCR. The fold change levels were highest in the CTDP1 (18q23), HDAC10 (22q13.33), KCNQ1 (11p15.5-p15.4), NINJ2 (12p13.33), NOTCH2 (1p12-p11.2), PCSK6 (15q26.3), SDHA (5p15.33), and MUC17 (7q22.1) genes.
CONCLUSION
Many candidate chromosomal regions that might be related to the pathogenesis of VA were detected by array CGH and should be systematically investigated to establish the causative and specific genes for VA.
MeSH Terms
Figure
Reference
-
1. Nakamura M, Takeshita A, Nose Y. Clinical characteristics associated with myocardial infarction, arrhythmias, and sudden death in patients with vasospastic angina. Circulation. 1987. 75:1110–1116.2. Pristipino YC, Beltrame JF, Finochiaro ML, et al. Major racial differences in coronary constrictor response between Japanese and Caucasians with recent myocardial infarction. Circulation. 2000. 101:1102–1108.3. Bertrand ME, LaBlanche JM, Tilmant PY, et al. Frequency of provoked coronary arterial spasm in 1089 consecutive patients undergoing coronary arteriography. Circulation. 1982. 65:1299–1306.4. Hibino H, Kurachi Y. A new insight into the pathogenesis of coronary vasospasm. Circ Res. 2006. 98:579–581.5. Chutkow WA, Pu J, Wheeler MT, et al. Episodic coronary artery vasospasm and hypertension develop in the absence of Sur2KATP channels. J Clin Invest. 2002. 110:203–208.6. Chang K, Baek SH, Seung KB, et al. The Glu298Asp polymorphism in the endothelial nitric oxide synthase gene is strongly associated with coronary spasm. Coron Artery Dis. 2003. 14:293–299.7. Park JS, Zhang SY, Jo SH, et al. Common adrenergic receptor polymorphisms as novel risk factors for vasospastic angina. Am Heart J. 2006. 151:864–869.8. Cho YS, Choi JH, Zhang SY, et al. Relationship of polymorphisms in the oxidative stress related genes: paraoxonase and p22phox - to variant angina and coronary artery stenosis in Korean. Korean Circ J. 2003. 33:104–112.9. Shimokawa H, Seto M, Katsumata N, et al. Rho-kinase-mediated pathway induces enhanced myosin light chain phosphorylations in a swine model of coronary artery spasm. Cardiovasc Res. 1999. 43:1029–1039.10. Ishida T, Hirata K, Sakoda T, et al. 5-HT1Dβ receptor mediates the supersensitivity of isolated coronary artery to serotonin in variant angina. Chest. 1998. 113:243–244.11. Shimizu M, Hata K, Takaoka H, et al. Sumatriptan provokes coronary artery spasm in patients with variant angina: possible involvement of serotonin 1B receptor. Int J Cardiol. 2007. 114:188–194.12. Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998. 20:207–211.13. Mantripragada KK, Buckley PG, Diaz de Ståhl T, Dumanski JP. Genomic microarrays in the spotlight. Trends Genet. 2004. 20:87–94.14. Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005. 6:331–354.15. Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001. 29:263–264.16. Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004. 36:299–303.17. Moon HJ, Yim SV, Lee KW, et al. Identification of DNA copy-number aberrations by array-comparative genomic hybridization in patients with schizophrenia. Biochem Biophys Res Commun. 2006. 344:531–539.18. Kim HS, Yim SV, Jung KH, et al. Altered DNA copy number in patients with different seizure disorder type: by array-CGH. Brain Dev. 2007. 29:639–643.19. Bonaglia MC, Giorda R, Tenconi R, et al. A 2.3Mb duplication of chromosome 8q24.3 associated with severe mental retardation and epilepsy detected by standard karyotype. Eur J Hum Genet. 2005. 13:586–591.20. Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005. 37:S11–S17.21. Choi YW, Choi JS, Zheng LT, et al. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the Lung. Lung Cancer. 2007. 55:43–51.22. Choi JS, Zheng LT, Ha E, et al. Comparative genomic hybridization array analysis and real-time PCR reveals genomic copy number alteration for lung adenocarcinomas. Lung. 2006. 184:355–362.23. Hurst CD, Fiegler H, Carr P, Williams S, Carter NP, Knowles MA. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004. 23:2250–2263.24. Mao X, Orchard G, Lillington DM, Russell-Jones R, Young BD, Whittaker S. Genetic alterations in primary cutaneous CD30+ anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2003. 37:176–185.25. Guillaud-Bataille M, Valent A, Soularue P, et al. Detecting single DNA copy number variations in complex genomes using one nanogram of starting DNA and BAC-array CGH. Nucleic Acids Res. 2004. 32:e112.26. Li Y, Zhang R, Qiao H, et al. Generation of insulin-producing cells from PDX1 gene-modified human esenchymal stem cells. J Cell Physiol. 2007. 211:36–44.27. Kang SM, Chang W, Lim S, et al. Losartan inhibits proliferation and inflammation of vascular smooth muscle cells by modulation of uric acid transporter. Tissue Eng Regen Med. 2008. 5:521–527.28. Wang G, Brennan C, Rook M, et al. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004. 32:e76.29. Calvo R, West J, Franklin W, et al. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci U S A. 2000. 97:12776–12781.30. Chujo M, Noguchi T, Miura T, Arinaga M, Uchida Y, Tagawa Y. Comparative genomic hybridization analysis detected frequent overrepresentation of chromosome 3q in squamous cell carcinoma of the lung. Lung Cancer. 2002. 38:23–29.31. Jeon JP, Shim SM, Jung JS, et al. A comprehensive profile of DNA copy number variations in a Korean population: identification of copy number invariant regions among Koreans. Exp Mol Med. 2009. 41:618–628.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Array-based Comparative Genomic Hybridization and Its Application to Cancer Genomes and Human Genetics
- Next generation sequencing and array-based comparative genomic hybridization for molecular diagnosis of pediatric endocrine disorders
- RAN-aCGH: R GUI Tools for Analysis and Visualization of an Array-CGH Experiment
- Analysis of Chromosomal Aberrations in Lung Cancer Cell Line, NCI-H1373
- Current Status and Future Clinical Applications of Array.based Comparative Genomic Hybridization