J Pathol Transl Med.
2016 Mar;50(2):104-112. 10.4132/jptm.2016.02.08.
Upregulated Neuro-oncological Ventral Antigen 1 (NOVA1) Expression Is Specific to Mature and Immature T- and NK-Cell Lymphomas
- Affiliations
-
- 1Department of Pathology, Yonsei University College of Medicine, Seoul, Korea. soyoon@yuhs.ac
- 2Anatomic Pathology Reference Lab, Seegene Medical Foundation, Seoul, Korea.
- 3Department of Internal Medicine, Division of Hematology, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2211377
- DOI: http://doi.org/10.4132/jptm.2016.02.08
Abstract
- BACKGROUND
Recent studies have revealed that the splicing factor neuro-oncological ventral antigen 1 (NOVA1) is enriched in fibroblasts and accumulated T cells of tertiary lymphoid structures. In the present study, we investigated NOVA1 expression in various subtypes of mature and immature T- and natural killer (NK)-cell lymphomas as well as in various B-cell lymphoma subtypes.
METHODS
NOVA1 immunoexpression was evaluated in hyperplastic palatine tonsils (n = 20), T- and NK-cell lymphomas (n = 177), diffuse large B-cell lymphomas (n = 151), and other types of B cell lymphomas (n = 31). Nuclear staining intensity and percentage of positive tumor cells were graded. NOVA1 mRNA expression was analyzed in various lymphoma cell lines.
RESULTS
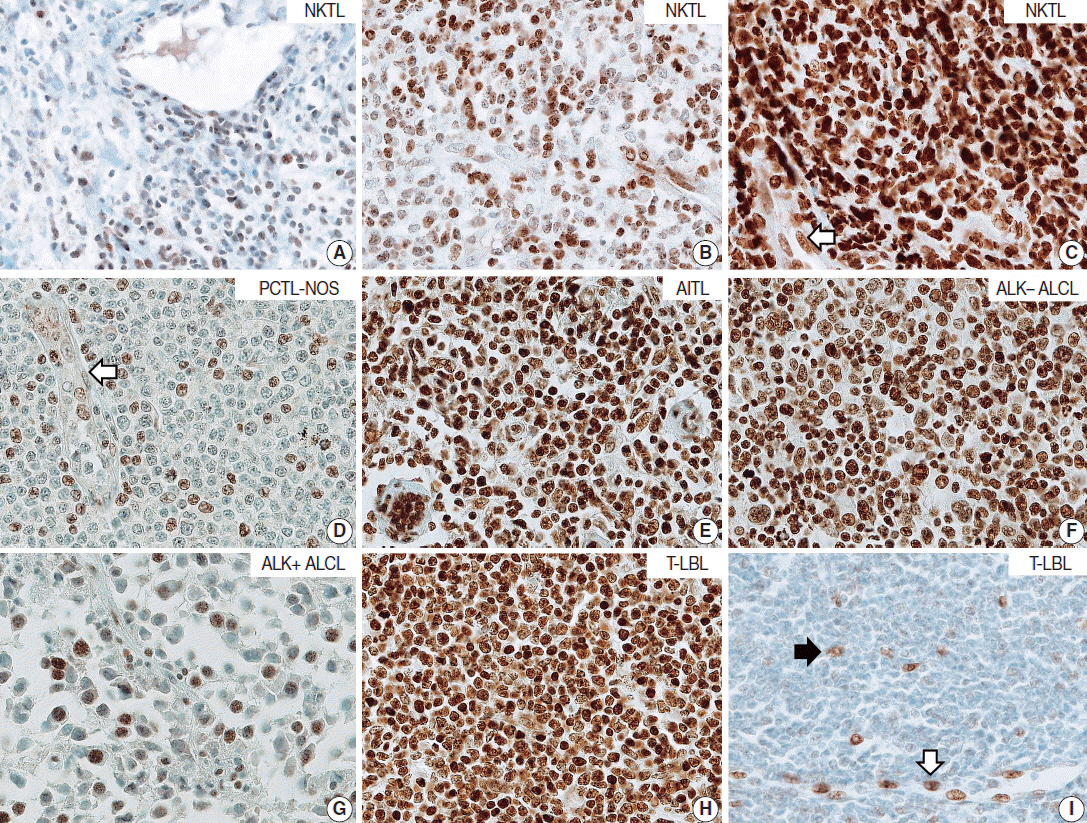
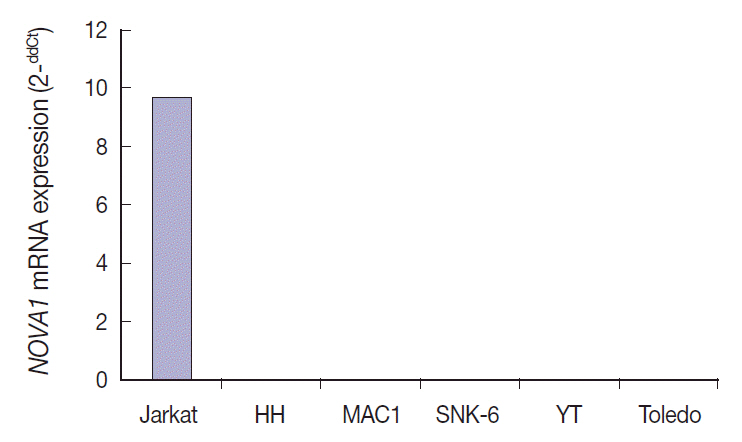
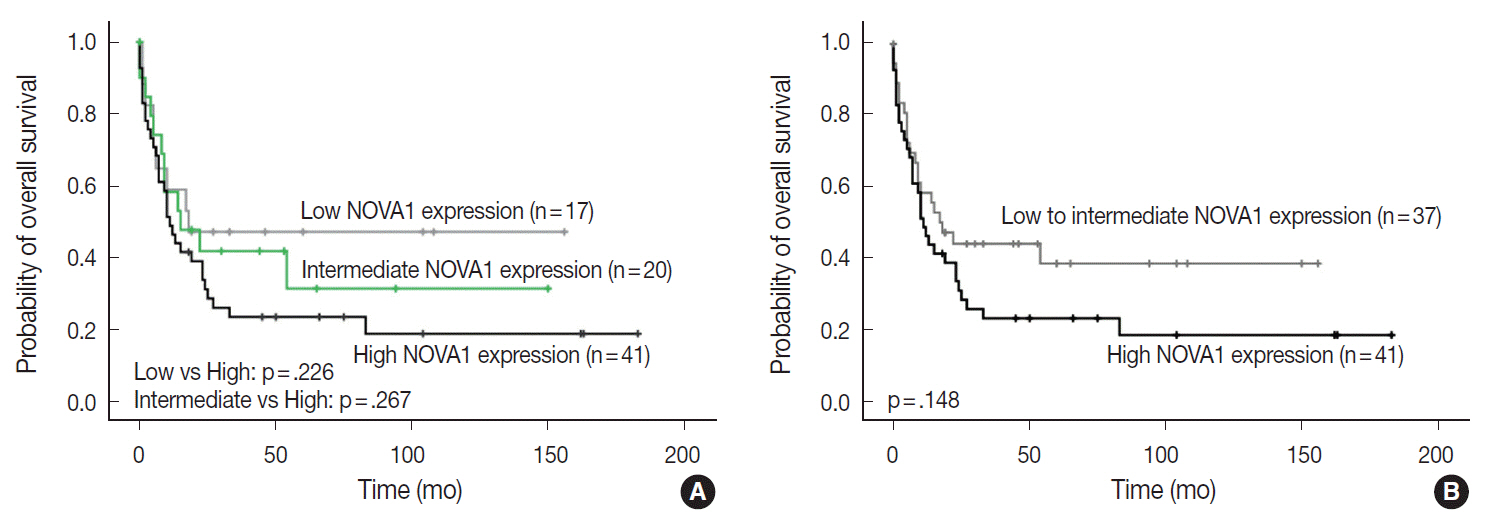
Tumor cells of T- and NK-cell lymphomas showed higher expression levels of NOVA1 than did normal paracortical T cells, and 56.5% of T- and NK-cell lymphoma cases showed diffuse and strong expression. The NOVA1 expression level varied according to the subtype; it was higher in angioimmunoblastic T-cell lymphoma, anaplastic lymphoma kinase (ALK)-negative anaplastic large cell lymphoma (ALCL), and T lymphoblastic leukemia/lymphoma (T-LBL), but it was lower in ALK-positive ALCL. In almost all B-cell lymphomas, NOVA1 expression was very low or negative. NOVA1 mRNA was also expressed in Jurkat, a T-LBL cell line.
CONCLUSIONS
The present findings suggest that NOVA1 upregulation may be involved in certain subtypes of T- and NK-cell lymphomas, but not in B-cell lymphomas. Upregulated NOVA1 expression seems to be a specific biological feature of activated T cells such as T- and NK-cell lymphomas.
Keyword
MeSH Terms
Figure
Reference
-
1. Barash Y, Calarco JA, Gao W, et al. Deciphering the splicing code. Nature. 2010; 465:53–9.
Article2. de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010; 38:2825–38.
Article3. Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012; 338:1593–9.
Article4. Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011; 12:715–29.
Article5. Mallinjoud P, Villemin JP, Mortada H, et al. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res. 2014; 24:511–21.
Article6. Yoon SO, Kim EK, Lee M, et al. NOVA1 inhibition by miR-146b-5p in the remnant tissue microenvironment defines occult residual disease after gastric cancer removal. Oncotarget. 2016; 7:2475–95.
Article7. Drayton DL, Liao S, Mounzer RH, Ruddle NH. Lymphoid organ development: from ontogeny to neogenesis. Nat Immunol. 2006; 7:344–53.
Article8. Ruddle NH, Akirav EM. Secondary lymphoid organs: responding to genetic and environmental cues in ontogeny and the immune response. J Immunol. 2009; 183:2205–12.
Article9. Buckley CD, Barone F, Nayar S, Bénézech C, Caamaño J. Stromal cells in chronic inflammation and tertiary lymphoid organ formation. Annu Rev Immunol. 2015; 33:715–45.
Article10. Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006; 6:392–401.
Article11. Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: IARC Press;2008.12. Park E, Park SY, Kim H, et al. Membranous insulin-like growth factor-1 receptor (IGF1R) expression is predictive of poor prognosis in patients with epidermal growth factor receptor (EGFR)-mutant lung adenocarcinoma. J Pathol Transl Med. 2015; 49:382–8.13. Falini B, Nicoletti I, Bolli N, et al. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007; 92:519–32.
Article14. Kelemen O, Convertini P, Zhang Z, et al. Function of alternative splicing. Gene. 2013; 514:1–30.
Article15. Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008; 321:686–91.
Article16. O’Rourke JP, Ness SA. Alternative RNA splicing produces multiple forms of c-Myb with unique transcriptional activities. Mol Cell Biol. 2008; 28:2091–101.
Article17. Henry C, Deschamps M, Rohrlich PS, et al. Identification of an alternative CD20 transcript variant in B-cell malignancies coding for a novel protein associated to rituximab resistance. Blood. 2010; 115:2420–9.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Aggressive NK/T-cell Lymphoma/Leukemia with Cutaneous Involvement in Adolescence
- Immunoreactivity of CD99 in Non-Hodgkin's Lymphoma: Unexpected Frequent Expression in ALK-positive Anaplastic Large Cell Lymphoma
- A Case of Nasal CD56+ NK/T Cell Lymphoma Mimicking Cellulitis which Developed after Persistent Orbital Swelling
- Basic immunohistochemistry for lymphoma diagnosis
- Therapeutic Outcome of Epstein-Barr Virus Positive T/NK Cell Lymphoma in the Upper Aerodigestive Tract