J Korean Acad Conserv Dent.
2011 Nov;36(6):498-505. 10.5395/JKACD.2011.36.6.498.
Microbial profile of asymptomatic and symptomatic teeth with primary endodontic infections by pyrosequencing
- Affiliations
-
- 1Department of Conservative Dentistry, Seoul National University School of Dentistry and Dental Research Institute, Seoul, Korea. kum6139@snu.ac.kr
- 2Department of Civil and Environmental Engineering, Yonsei University, Seoul, Korea.
- 3Department of Dentistry, Yonsei University Wonju College of Medicine, Wonju, Korea.
- KMID: 2176604
- DOI: http://doi.org/10.5395/JKACD.2011.36.6.498
Abstract
OBJECTIVES
The purpose of this in vivo study was to investigate the microbial diversity in symptomatic and asymptomatic canals with primary endodontic infections by using GS FLX Titanium pyrosequencing.
MATERIALS AND METHODS
Sequencing was performed on 6 teeth (symptomatic, n = 3; asymptomatic, n = 3) with primary endodontic infections. Amplicons from hypervariable region of the small-subunit ribosomal RNA gene were generated by polymerized chain reaction (PCR), and sequenced by means of the GS FLX Titanium pyrosequencing.
RESULTS
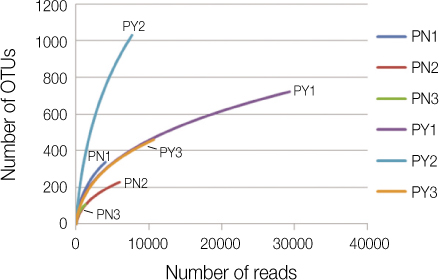
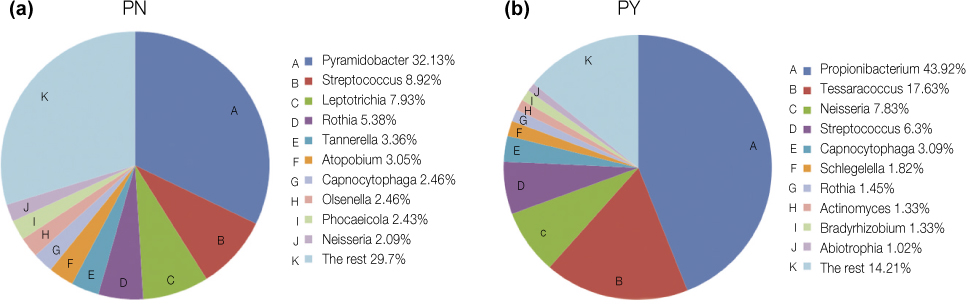
On average, 10,639 and 45,455 16S rRNA sequences for asymptomatic and symptomatic teeth were obtained, respectively. Based on Ribosomal Database Project Classifier analysis, pyrosequencing identified the 141 bacterial genera in 13 phyla. The vast majority of sequences belonged to one of the seven phyla: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Spirochetes, and Synergistetes. In genus level, Pyramidobacter, Streptococcus, and Leptotrichia constituted about 50% of microbial profile in asymptomatic teeth, whereas Neisseria, Propionibacterium, and Tessaracoccus were frequently found in symptomatic teeth (69%). Grouping the sequences in operational taxonomic units (3%) yielded 450 and 1,997 species level phylotypes in asymptomatic and symptomatic teeth, respectively. The total bacteria counts were significantly higher in symptomatic teeth than that of asymptomatic teeth (p < 0.05).
CONCLUSIONS
GS FLX Titanium pyrosequencing could reveal a previously unidentified high bacterial diversity in primary endodontic infections.
Keyword
MeSH Terms
Figure
Reference
-
1. Kakehashi S, Stanley HR, Fitzgerald RJ. The effects of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral Surg Oral Med Oral Pathol. 1965. 20:340–349.
Article2. Gomes BP, Drucker DB, Lilley JD. Associations of specific bacteria with some endodontic signs and symptoms. Int Endod J. 1994. 27:291–298.
Article3. Gomes BP, Lilley JD, Drucker DB. Associations of endodontic symptoms and signs with particular combinations of specific bactreria. Int Endod J. 1996. 29:69–75.
Article4. Yoshida M, Fukushima H, Yamamoto K, Ogawa K, Toda T, Sagawa H. Correlation between clinical symptoms and microorganism isolated from root canal of teeth with periapical pathosis. J Endod. 1987. 13:24–28.
Article5. Rôças IN, Siqueira JF Jr. Root canal microbiota of teeth with chronic apical periodontitis. J Clin Microbiol. 2008. 46:3599–3606.
Article6. Siqueira JF Jr, Roôças IN. Community as the unit of pathogenicity: an emerging concept as to the microbial pathogenesis of apical periodontitis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009. 107:870–878.
Article7. Kim SY, Choi HY, Park SH, Choi GW. Distribution of oral pathogens in infection of endodontic origin. J Korean Acad Conserv Dent. 2003. 28:303–313.
Article8. Kum KY, Fouad AF. PCR-based identification of Eubacteirum species in endodontic infection. J Korean Acad Conserv Dent. 2003. 28:241–248.
Article9. Lee YJ, Kim MK, Hwang HK, Kook JK. Isolation and identification of bacteria from the root canal of the teeth diagnosed as the acute pulpitis and acute periapical abscess. J Korean Acad Conserv Dent. 2005. 30:409–422.
Article10. Siqueira JF Jr, Rôças IN. Diversity of endodontic microbiota revisited. J Dent Res. 2009. 88:969–981.
Article11. Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008. 87:1016–1020.
Article12. Horz HP, Vianna ME, Gomes BP, Conrads G. Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J Clin Microbiol. 2005. 43:5332–5337.
Article13. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplication for phylogenetic study. J Bacteriol. 1991. 173:697–703.
Article14. Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007. 8:R143.
Article15. Li L, Hsiao WW, Nandakumar R, Barbuto SM, Mongodin EF, Paster BJ, Fraser-Liggett CM, Fouad AF. Analyzing endodontic infections by deep coverage pyrosequencing. J Dent Res. 2010. 89:980–984.
Article16. Lee TK, Van Doan T, Yoo K, Choi S, Kim C, Park J. Discovery of commonly existing anode biofilm microbes in two different wastewater treatment MFCs using FLX Titanium pyrosequencing. Appl Microbiol Biotechnol. 2010. 87:2335–2343.
Article17. Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, Russ C, Lander ES, Nusbaum C, Jaffe DB. Quality scores and SNP detection in sequencing-by-synthesis system. Genome Res. 2008. 18:763–770.18. Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007. 57:2259–2261.
Article19. Siqueira JF Jr, Rôças IN, Paiva SS, Magalhães KM, Guimarães-Pinto T. Cultivable bacteria in infected root canals as identified by 16S rRNA gene sequencing. Oral Microbiol Immunol. 2007. 22:266–271.
Article20. Vickerman MM, Brossard KA, Funk DB, Jesionowski AM, Gill SR. Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J Med Microbiol. 2007. 56:110–118.
Article21. Siqueira JF Jr, Alves FR, Rôças IN. Pyrosequencing analysis of the apical root canal microbiota. J Endod. 2011. 37:1499–1503.
Article22. Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009. 9:259.
Article23. Lazarevic V, Whiteson K, Hernandez D, François P, Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics. 2010. 11:523.
Article24. Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osterås M, Schrenzel J, François P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009. 79:266–271.
Article25. Nercessian O, Fouquet Y, Pierre C, Prieur D, Jeanthon C. Diversity of Bacteria and Archaea associated with a carbonate-rich metalliferous sediment sample from the Rainbow vent field on the Mid-Atlantic Ridge. Environ Microbiol. 2005. 7:698–714.
Article26. Signoretto C, Burlacchini G, Bianchi F, Cavalleri G, Canepari P. Differences in microbiological composition of saliva and dental plaque in subjects with different drinking habits. New Microbiol. 2006. 29:293–302.27. Sundqvist G, Figdor D. Life as an endodontic pathogen: ecological differences between the untreated and root filled root canals. Endod Top. 2003. 6:3–28.
Article28. Rôças IN, Siqueira JF Jr, Debelian GJ. Analysis of symptomatic and asymptomatic primary root canal infections in adult Norwegian patients. J Endod. 2011. 37:1206–1212.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Distribution of oral pathogens in infections of endodontic origin
- Prevalence of referral reasons and clinical symptoms for endodontic referrals
- Analysis of Amelogenin Gene & Short Tandem Repeat(STR) Locus F13A01, LPL from Pulpless Teeth Dentin
- Bacterial diversity and its relationship to growth performance of broilers
- Diagnosis and treatment of teeth with primary endodontic lesions mimicking periodontal disease: three cases with long-term follow ups