Blood Res.
2013 Dec;48(4):242-249. 10.5045/br.2013.48.4.242.
Next generation sequencing: new tools in immunology and hematology
- Affiliations
-
- 1Department of Life and Reproduction Science, University of Verona, Verona, Italy.
- 2Hematology Unit, Bolzano Central Hospital, Bolzano, Italy.
- 3Department of Pathology and Diagnostics, University of Verona, Verona, Italy. vladia.monsurro@univr.it
- KMID: 2172870
- DOI: http://doi.org/10.5045/br.2013.48.4.242
Abstract
- One of the hallmarks of the adaptive immune system is the specificity of B and T cell receptors. Thanks to somatic recombination, a large repertoire of receptors can be generated within an individual that guarantee the recognition of a vast number of antigens. Monoclonal antibodies have limited applicability, given the high degree of diversity among these receptors, in BCR and TCR monitoring. Furthermore, with regard to cancer, better characterization of complex genomes and the ability to monitor tumor-specific cryptic mutations or translocations are needed to develop better tailored therapies. Novel technologies, by enhancing the ability of BCR and TCR monitoring, can help in the search for minimal residual disease during hematological malignancy diagnosis and follow-up, and can aid in improving bone marrow transplantation techniques. Recently, a novel technology known as next generation sequencing has been developed; this allows the recognition of unique sequences and provides depth of coverage, heterogeneity, and accuracy of sequencing. This provides a powerful tool that, along with microarray analysis for gene expression, may become integral in resolving the remaining key problems in hematology. This review describes the state of the art of this novel technology, its application in the immunological and hematological fields, and the possible benefits it will provide for the hematology and immunology community.
MeSH Terms
-
Allergy and Immunology*
Antibodies, Monoclonal
Bone Marrow Transplantation
Diagnosis
Gene Expression
Genome
Hematologic Neoplasms
Hematology*
Immune System
Microarray Analysis
Monitoring, Immunologic
Neoplasm, Residual
Population Characteristics
Receptors, Antigen, T-Cell
Recombination, Genetic
Sensitivity and Specificity
Antibodies, Monoclonal
Receptors, Antigen, T-Cell
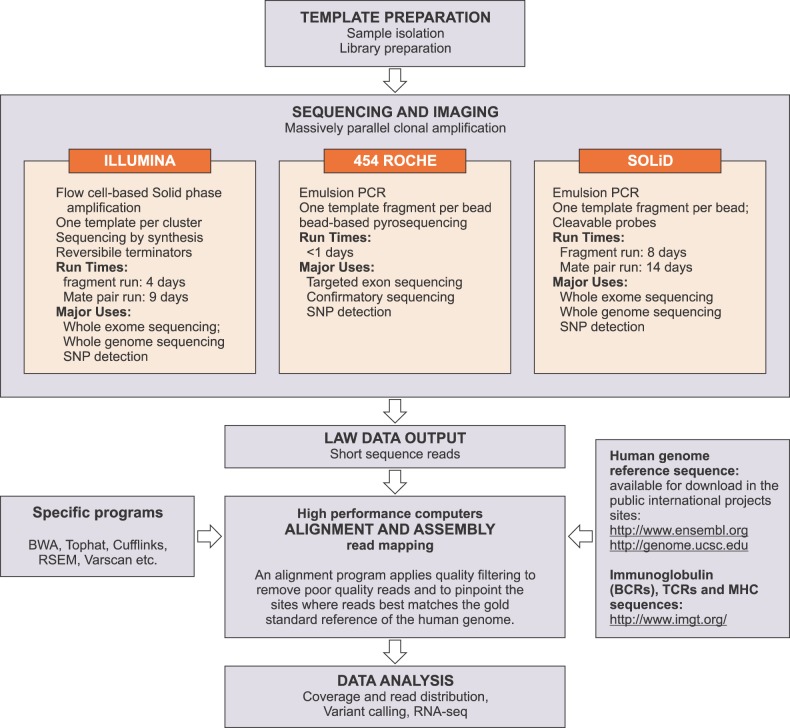
Figure
Cited by 2 articles
-
Favorable long-term survival using consolidation chemotherapy without allogeneic hematopoietic cell transplantation for acute myeloid leukemia with wild-type NPM1 without FLT3-ITD
Dong Won Baek, Jung Min Lee, Ju-Hyung Kim, Hee Jeong Cho, Ji-Yeon Ham, Jang-Soo Suh, Sang-Kyun Sohn, Joon Ho Moon
Blood Res. 2019;54(3):189-197. doi: 10.5045/br.2019.54.3.189.Efficacy of Annexin A1 Immunostaining in Bone Marrow for the Diagnosis of Hairy Cell Leukemia
Chang-Hun Park, Hyun-Young Kim, Sang-Yong Shin, Hee-Jin Kim, Chul Won Jung, Jong-Won Kim, Sun-Hee Kim
Lab Med Online. 2019;9(4):236-241. doi: 10.3343/lmo.2019.9.4.236.
Reference
-
1. Tonegawa S, Steinberg C, Dube S, Bernardini A. Evidence for somatic generation of antibody diversity. Proc Natl Acad Sci U S A. 1974; 71:4027–4031. PMID: 4215074.
Article2. Tonegawa S. Somatic generation of antibody diversity. Nature. 1983; 302:575–581. PMID: 6300689.
Article3. Kim S, Davis M, Sinn E, Patten P, Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981; 27:573–581. PMID: 6101208.
Article4. Thomas L. On immunosurveillance in human cancer. Yale J Biol Med. 1982; 55:329–333. PMID: 6758376.5. Burnet FM. Immunological aspects of malignant disease. Lancet. 1967; 1:1171–1174. PMID: 4165129.
Article6. Monsurro V, Nagorsen D, Wang E, et al. Functional heterogeneity of vaccine-induced CD8(+) T cells. J Immunol. 2002; 168:5933–5942. PMID: 12023400.
Article7. Monsurro V, Wang E, Yamano Y, et al. Quiescent phenotype of tumor-specific CD8+ T cells following immunization. Blood. 2004; 104:1970–1978. PMID: 15187028.8. Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011; 52:413–435. PMID: 21698376.
Article9. Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010; 11:31–46. PMID: 19997069.
Article10. Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011; 9:34. PMID: 21627854.
Article11. Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011; 8:469–477. PMID: 21623353.
Article12. Bowers J, Mitchell J, Beer E, et al. Virtual terminator nucleotides for next-generation DNA sequencing. Nat Methods. 2009; 6:593–595. PMID: 19620973.
Article13. Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009; 19:1817–1824. PMID: 19541912.
Article14. Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009; 114:4099–4107. PMID: 19706884.15. Robins HS, Srivastava SK, Campregher PV, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010; 2:47ra64.16. Venturi V, Quigley MF, Greenaway HY, et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J Immunol. 2011; 186:4285–4294. PMID: 21383244.
Article17. Warren RL, Freeman JD, Zeng T, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011; 21:790–797. PMID: 21349924.
Article18. Jiang N, Weinstein JA, Penland L, White RA 3rd, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci U S A. 2011; 108:5348–5353. PMID: 21393572.
Article19. Boyd SD, Marshall EL, Merker JD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009; 1:12ra23.
Article20. Boyd SD, Gaeta BA, Jackson KJ, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010; 184:6986–6992. PMID: 20495067.
Article21. Glanville J, Zhai W, Berka J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009; 106:20216–20221. PMID: 19875695.
Article22. Ge X, Mazor Y, Hunicke-Smith SP, Ellington AD, Georgiou G. Rapid construction and characterization of synthetic antibody libraries without DNA amplification. Biotechnol Bioeng. 2010; 106:347–357. PMID: 20198660.
Article23. Reddy ST, Ge X, Miklos AE, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010; 28:965–969. PMID: 20802495.
Article24. Zhai W, Glanville J, Fuhrmann M, et al. Synthetic antibodies designed on natural sequence landscapes. J Mol Biol. 2011; 412:55–71. PMID: 21787786.
Article25. Nguyen P, Ma J, Pei D, Obert C, Cheng C, Geiger TL. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire. BMC Genomics. 2011; 12:106. PMID: 21310087.
Article26. Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012; 135:183–191. PMID: 22043864.
Article27. Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002; 346:1937–1947. PMID: 12075054.28. Dohner K, Dohner H. Molecular characterization of acute myeloid leukemia. Haematologica. 2008; 93:976–982. PMID: 18591623.
Article29. Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012; 44:1321–1325. PMID: 23143597.
Article30. Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010; 362:1417–1429. PMID: 20393178.
Article31. Kohlmann A, Grossmann V, Nadarajah N, Haferlach T. Next-generation sequencing - feasibility and practicality in haematology. Br J Haematol. 2013; 160:736–753. PMID: 23294427.
Article32. Arber DA, Brunning RD, Le Beau MM, et al. Acute myeloid leukaemia with recurrent genetic abnormalities. In : Swerdlow SH, Campo E, Harris NL, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon, France: IARC;2008. p. 110–123.33. Walter RB, Othus M, Burnett AK, et al. Significance of FAB subclassification of "acute myeloid leukemia, NOS" in the 2008 WHO classification: analysis of 5848 newly diagnosed patients. Blood. 2013; 121:2424–2431. PMID: 23325837.
Article34. Welch JS, Link DC. Genomics of AML: clinical applications of next-generation sequencing. Hematology Am Soc Hematol Educ Program. 2011; 2011:30–35. PMID: 22160009.
Article35. Logan AC, Gao H, Wang C, et al. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc Natl Acad Sci U S A. 2011; 108:21194–21199. PMID: 22160699.
Article36. Wu D, Sherwood A, Fromm JR, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med. 2012; 4:134ra63.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analytical Tools and Databases for Metagenomics in the Next-Generation Sequencing Era
- Molecular Risk Stratification using Next-generation Sequencing in Acute Myeloid Leukemia
- Recent Advances in the Clinical Application of Next-Generation Sequencing
- Accelerating next generation sequencing data analysis: an evaluation of optimized best practices for Genome Analysis Toolkit algorithms
- Utility of Next-Generation Sequencing for Deciphering Intratumor Heterogeneity in Prostate Cancer