J Bacteriol Virol.
2015 Dec;45(4):285-303. 10.4167/jbv.2015.45.4.285.
MinION(TM): New, Long Read, Portable Nucleic Acid Sequencing Device
- Affiliations
-
- 1Department of Microbiology, Chung Ang University, College of Medicine, Seoul 06974, Korea. kimwy@cau.ac.kr
- 2School of Biology, Newcastle University, Newcastle upon Tyne, NE1 7RU, UK.
- KMID: 2168713
- DOI: http://doi.org/10.4167/jbv.2015.45.4.285
Abstract
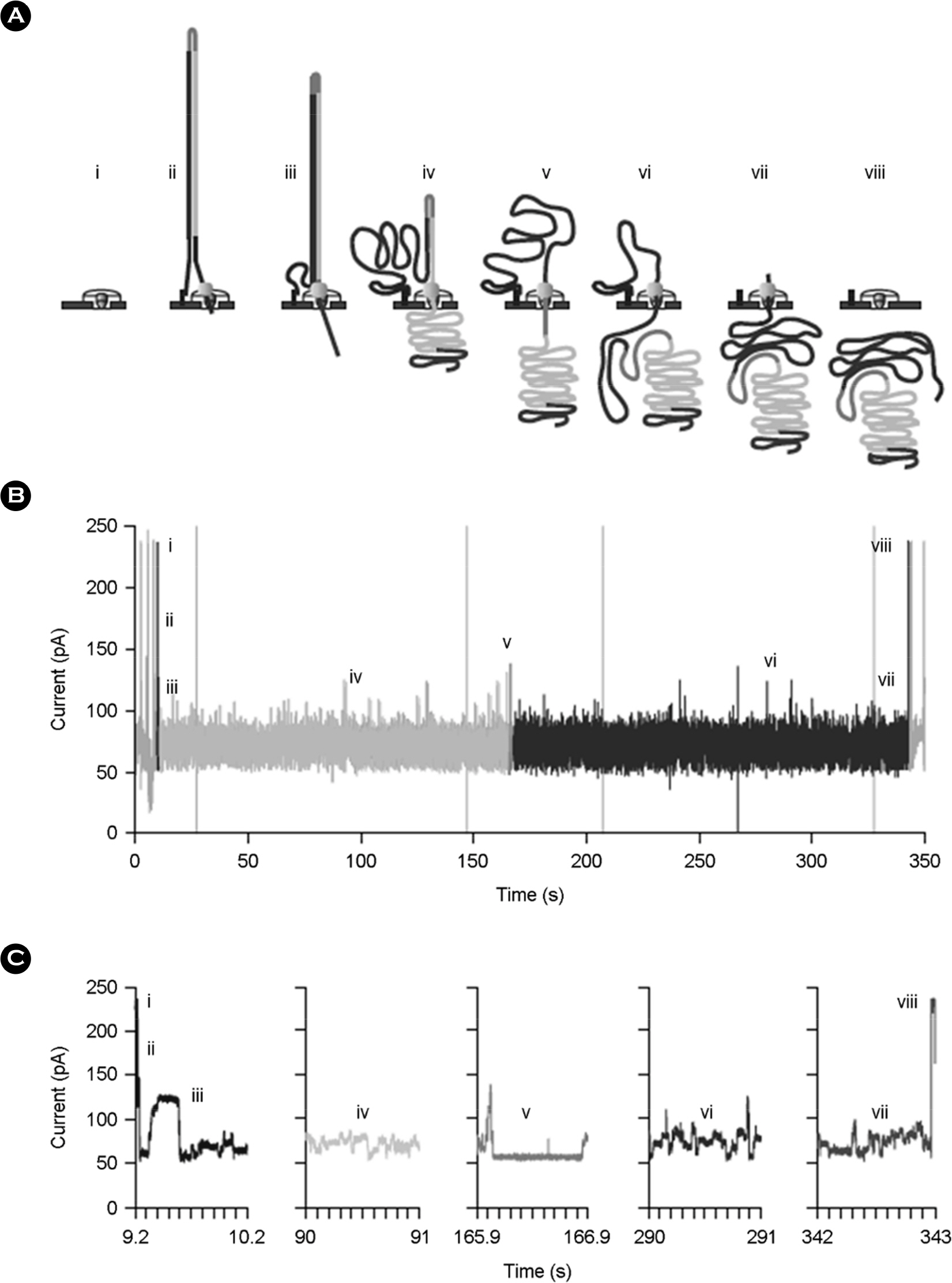
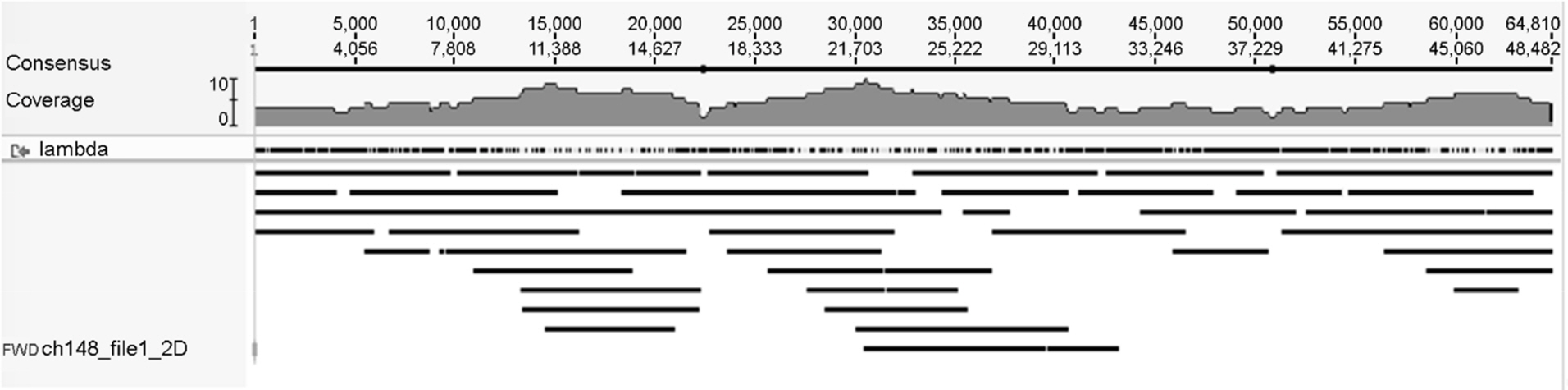
- The MinION(TM) is a miniature nanopore-based analysis device in which the characteristics of an analyte, as it passes through the nanopore, cause changes in the flow of ions through the pore, which are measured, as current flow, by a low noise amplifier and analogue-to-digital converter. Potentially any molecular analyte capable of passing through the nanopore may modify the flow of ions and generate a signal which might be diagnostic. In practice the current device is focussed on DNA sequencing, directly sequencing RNA is a likely development. With the MinION Access Program making the MinION(TM) widely available a flood of applications exploiting its real time, long read capabilities have been published. We review the background to the technology and compare it to current next generation sequencing.
Keyword
Figure
Reference
-
1). Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Human Mol Genet. 2010; 19:R227–40.
Article2). Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977; 74:5463–7.
Article3). Glenn TC. Field Guide to Next Generation DNA Sequencers. Mol Ecol Resour. 2011; 11:759–69.4). Glenn TC. http://www.molecularecologist.com/next-gen-fieldguide-2014/. (accessed July 2015). 2014.5). Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The Next-Generation Sequencing Revolution and Its Impact on Genomics. Cell. 2013; 155:27–38.
Article6). Morey M, Fernández-Marmiesse A, Castiñeiras D, Fraga JM, Couce ML, Cocho JA. A glimpse into past, present, and future DNA sequencing. Mol Genet Metab. 2013; 110:3–24.
Article7). Smith MI, Turpin W, Tyler AD, Silverberg MS, Croitoru K. Microbiome analysis – from technical advances to biological relevance. F1000Prime Rep. 2014; 6:51.
Article8). Erlich Y, Mitra PP, de la Bastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat Methods. 2008; 5:679–82.
Article9). Rusk N. Torrents of sequence. Nature Methods. 2011; 8:44.
Article10). Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008; 456:53–9.11). Kawashima EH, Laurent F, Pascal M. Patent (2005-05-12): Method of nucleic acid amplification.http://www.patentlens.net/patentlens/patent/WO_1998_044151_A1/en/. (accessed 22 July 2015). 2005.12). Williams R, Peisajovich SG, Miller OJ, Magdassi S, Tawfik DS, Griffiths AD. Amplification of complex gene libraries by emulsion PCR. Nat Methods. 2006; 3:545–50.
Article13). Mardis ER. The $1, 000 genome, the $100, 000 analysis? Genome Med. 2010; 2:84.14). Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics. 2005; 21:4320–1.
Article15). Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, et al. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010; 329:533–8.16). Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bähler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012; 151:671–83.
Article17). Miura F, Kawaguchi N, Yoshida M, Uematsu C, Kito K, Sakaki Y, et al. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics. 2008; 9:574.
Article18). Liu Y, Ferguson JF, Xue C, Silverman IM, Gregory B, Reilly MP. Evaluating the Impact of Sequencing Depth on Transcriptome Profiling in Human Adipose. PLoS One. 2013; 8:e66883.
Article19). SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014; 32:903–14.20). Pretto DI, Eid JS, Yrigollen CM, Tang HT, Loomis EW, Raske C, et al. Differential increases of specific FMR1 mRNA isoforms in premutation carriers. J Med Genet. 2015; 52:42–52.21). Jaffrey SR. An expanding universe of mRNA modifications. Nat Struct Mol Biol. 2014; 21:945–6.
Article22). Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol. 2009; 75:5227–36.
Article23). Wooley JC, Godzik A, Friedberg I. A Primer on Metagenomics. PLoS Comput Biol. 2010; 6:e1000667.
Article24). Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, van Elsas JD, et al. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol. 2009; 7:252.
Article25). DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006; 72:5069–72.
Article26). Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO, et al. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol. 2008; 74:5422–8.
Article27). Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007; 17:377–86.
Article28). Wommack KE, Bhavsar J, Ravel J. Metagenomics: Read Length Matters. Appl Environ Microbiol. 2008; 74:453–63.
Article29). Moran AM. Metatranscriptomics: eavesdropping on complex microbial communities. Microbe. 2009; 4:329–35.
Article30). Carvalhais LC, Dennis PG, Tyson GW, Schenk PM. Application of metatranscriptomics to soil environments. J Microbiol Methods. 2012; 91:246–51.
Article31). Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003; 299:682–6.
Article32). Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-Time DNA sequencing from single polymerase molecules. Science. 2009; 323:133–8.
Article33). Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010; 464:1012–7.
Article34). McCarthy A. Third Generation DNA Sequencing: Pacific Biosciences' single molecule real time technology. Chem Biol. 2010; 17:675–6.
Article35). Reuter JA, Spacek DV, Snyder MP. High-Throughput Sequencing Technologies. Mol Cell. 2015; 58:586–97.
Article36). Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010; 38:e159.
Article37). English AC, Richards S, Han Y, Wang M, Vee V, Qu J, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012; 7:e47768.
Article38). Ribeiro FJ, Przybylski D, Yin S, Sharpe T, Gnerre S, Abouelleil A, et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 2012; 22:2270–7.
Article39). Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012; 30:693–700.40). Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, Mcvey SD, et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013; 14:R101.
Article41). Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015; 517:608–11.
Article42). Flusberg FA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010; 7:461–5.
Article43). Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012; 30:1232–9.44). Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013; 31:1009–14.
Article45). Tilgner H, Grubert F, Sharon D, Snyder MP. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci U S A. 2014; 111:9869–74.
Article46). Loman NJ, Watson M. Successful test launch for nanopore sequencing. Nat Methods. 2015; 12:303–4.
Article47). Deamer DW, Akeson M. Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol. 2000; 18:147–51.
Article48). Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc Chem Res. 2002; 35:817–25.
Article49). Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996; 93:13770–3.
Article50). Pennisi E. Genome sequencing. Search for pore-fection. Science. 2012; 336:534–7.51). Bayley H. Nanopore Sequencing: From imagination to reality. Clin Chem. 2015; 61:25–31.
Article52). Wanunu M. Nanopores: A journey towards DNA sequencing. Phys Life Rev. 2012; 9:125–58.
Article53). Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci U S A. 2008; 105:20647–52.
Article54). Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009; 4:765–72.
Article55). Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996; 274:1859–66.
Article56). Füssle R, Bhakdi S, Sziegoleit A, Tranum-Jensen J, Kranz T, Wellensiek HJ. On the mechanism of membrane damage by Staphylococcus aureus α-toxin. J Cell Biol. 1981; 91:83–94.57). Gouaux E. α-Hemolysin from Staphylococcus aureus: an archetype of β-barrel, channel-forming toxins. J Struct Biol. 1998; 21:110–22.58). Meller A, Branton D. Single molecule measurements of DNA transport through a nanopore. Electrophoresis. 2002; 23:2583–91.
Article59). Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999; 77:3227–33.
Article60). Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci U S A. 2000; 97:1079–84.
Article61). Kawano R, Schibel AE, Cauley C, White HS. Controlling the translocation of single-stranded DNA through alpha-hemolysin ion channels using viscosity. Langmuir. 2009; 25:1233–7.62). Mitchell N, Howorka S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew Chem Int Ed Engl. 2008; 47:5565–8.63). Zhang Y, Liu L, Sha J, Ni Z, Yi H, Chen Y. Nanopore detection of DNA molecules in magnesium chloride solutions. Nanoscale Res Lett. 2013; 8:245.
Article64). Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci U S A. 2008; 105:19720–5.65). Rincon-Restrepo M, Mikhailova E, Bayley H, Maglia G. Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes. Nano Lett. 2011; 11:746–50.
Article66). Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at Five Angstrom Precision. Nat Biotechnol. 2012; 30:344–8.67). Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J Am Chem Soc. 2010; 132:17961–72.
Article68). Stoddart D, Franceschini L, Heron A, Bayley H, Maglia G. DNA stretching and optimization of nucleobase recognition in enzymatic nanopore sequencing. Nanotechnology. 2015; 26:084002.
Article69). Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 2010; 5:798–806.
Article70). Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008; 26:1146–53.
Article71). Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci U S A. 2009; 106:7702–7.
Article72). Ervin EN, Barrall GA, Pal P, Bean MK, Schibel AE, Hibbs AD. Creating a single sensing zone within an alpha-hemolysin pore via site-directed mutagenesis. Bionanoscience. 2014; 4:78–84.
Article73). Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012; 30:349–53.
Article74). Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS One. 2011; 6:e25723.
Article75). Stoddart D, Heron AJ, Klingelhoefer J, Mikhailova E, Maglia G, Bayley H. Nucleobase recognition in ssDNA at the central constriction of the α-hemolysin pore. Nano Lett. 2010; 10:3633–7.
Article76). Stoddart D, Maglia G, Mikhailova E, Heron AJ, Bayley H. Multiple base-recognition sites in a biological nanopore: two heads are better than one. Angew Chem Int Ed Engl. 2010; 49:556–9.
Article77). Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun. 2010; 46:8195–7.
Article78). Schreiber J, Karplus K. Segmentation of noisy signals generated by a nanopore. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/014258.
Article79). Schreiber J, Karplus K. Analysis of nanopore data using Hidden Markov Models. Bioinformatics. 2015; 31:1897–903.
Article80). Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014; 32:829–33.
Article81). Timp W, Comer J, Aksimentiev A. DNA base-calling from a nanopore using a Viterbi algorithm. Biophys J. 2012; 102:L37–9.
Article82). Viterbi AJ. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. Inf Theory IEEE Trans. 1967; 3:260–9.
Article83). Urban JM, Bliss J, Lawrence CE, Gerbi SA. Sequencing ultra-long DNA molecules with the Oxford Nanopore MinION. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/019281.
Article84). Mikheyev AS, Tin MM. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour. 2014; 14:1097–102.
Article85). Madoui MA, Engelen S, Cruaud C, Belser C, Bertrand L, Alberti A, et al. Genome assembly using nanopore-guided long and error-free DNA reads. BMC Genomics. 2015; 16:327.
Article86). Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015; 12:733–5.87). Quick J, Quinlan AR, Loman NJ. A reference bacterial genome dataset generated on the MinION(TM) portable single-molecule nanopore sequencer. Gigascience. 2014; 3:22.
Article88). Evans TC. DNA Damage the major cause of missing pieces from the DNA puzzle. NEB Expressions. 2007; 2:1.89). Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. Improved data analysis for the MinION nanopore sequencer. Nat Methods. 2015; 12:351–6.
Article90). The HDF Group. Hierarchical Data Format, version 5. 1997–2015. http://www.hdfgroup.org/HDF5/. (accessed Aug 2015). 2015.91). Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics. 2014; 30:3399–401.
Article92). Watson M, Thomson M, Risse J, Talbot R, Santoyo-Lopez J, Gharbi K, et al. pore: an R package for the visualization and analysis of nanopore sequencing data. Bioinformatics. 2015; 31:114–5.
Article93). R Core Team. R: A Language and Environment for Statistical Computing.https://www.R-project.org. (accessed Aug 2015). 2015.94). Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011; 21:487–93.95). Sovic I, Sikic M, Wilm A, Fenlon SN, Chen S, Nagarajan N. Fast and sensitive mapping of error-prone nanopore sequencing reads with GraphMap. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/020719.
Article96). Warren RL, Yang C, Vandervalk BP, Behsaz B, Lagman A, Jones SJ, et al. LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience. 2015; 4:35.
Article97). Koren S, Phillippy AM. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol. 2015; 23:110–20.
Article98). Karlsson E, Lärkeryd A, Sjödin A, Forsman M, Stenberg P. Scaffolding of a bacterial genome using MinION nanopore sequencing. Sci Rep. 2015; 5:11996.
Article99). Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014; 15:211.
Article100). Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012; 13:238.
Article101). Risse J, Thomson M, Blakely G, Koutsovoulos G, Blaxter M, Watson M. A single chromosome assembly of Bacteroides fragilis strain BE1 from Illumina and MinION nanopore sequencing data. BioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/024323.
Article102). Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz M, McCombie WR. Oxford Nanopore Sequencing and de novo Assembly of a Eukaryotic Genome. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/013490.103). Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015; 33:296–300.
Article104). Ammar R, Paton TA, Torti D, Shlien A, Bader GD. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Res. 2015; 4:17.
Article105). Szalay T, Golovchenko JA. A de novo DNA Sequencing and Variant Calling Algorithm for Nanopores. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/019448.106). Ayub M, Hardwick SW, Luisi BF, Bayley H. Nanopore-Based Identification of Individual Nucleotides for Direct RNA Sequencing. Nano Lett. 2013; 13:6144–50.
Article107). Schreiber J, Wescoe ZL, Abu-Shumays R, Vivian JT, Baatar B, Karplus K, et al. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc Natl Acad Sci U S A. 2013; 110:18910–5.
Article108). Bolisetty M, Rajadinakaran G, Graveley B. Determining Exon Connectivity in Complex mRNAs by Nanopore Sequencing. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/019752.
Article109). Benitez-Paez A, Portune K, Sanz Y. Species level resolution of 16S rRNA gene amplicons sequenced through MinION TM portable nanopore sequencer. Bio-Rxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/021758.110). Cao MD, Ganesamoorthy D, Elliott A, Zhang H, Cooper M, Coin L. Real-time strain typing and analysis of antibiotic resistance potential using Nanopore Min-ION sequencing. BioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/019356.111). Quick J, Ashton P, Calus S, Chatt C, Gossain S, Hawker J, et al. Rapid draft sequencing and real-time nanopore sequencing in a hospital out break of Salmonella. Genome Biol. 2015; 16:114.
Article112). Wang J, Moore NE, Deng YM, Eccles DA, Hall RJ. MinION nanopore sequencing of an influenza genome. Front Microbiol. 2015; 6:766.
Article113). Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. bioRxiv. 2015. DOI: doi: http://dx.doi.org/10.1101/020420.
Article114). Kilianski A, Haas JL, Corriveau EJ, Liem AT, Willis KL, Kadavy DR, et al. Bacterial and viral identification and differentiation by amplicon sequencing on the MinION nanopore sequencer. Gigascience. 2015; 4:12.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Whole Genome Analysis of SARSCoV-2 Using Oxford Nanopore MinION System
- Long-read sequencing of 12 samples discovered novel variants within human leukocyte antigen region
- Long-read next-generation sequencing for molecular diagnosis of pediatric endocrine disorders
- Storing Digital Information in Long-Read DNA
- Amplicon-Based MinION Sequencing Complements Severe Fever With Thrombocytopenia Syndrome (SFTS) Diagnosis via Real-Time RT-PCR in Patients With Suspected SFTS